For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFGQLYDRALDGERCWVRHDDGRVKNLPVRRWLGGSGGDATFDHAVVGLCHGPTIDLGCGPGRLVAHLVQ RGVPALGVDLSATAVELARRSGAPALRRDVFEPLPGTGRWQTVLLADGNVGLGGDPRRVLGRATELLRRG GRCLAELDGTTSGVDVGWVRLESHRMIGPWFRWASVGIDCVPSLAAEAGLAVGGIHRIGTRVVADLVA
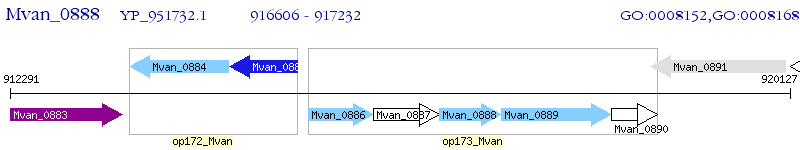
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0888 | - | - | 100% (208) | methyltransferase type 12 |
| M. vanbaalenii PYR-1 | Mvan_1888 | - | 1e-05 | 38.33% (60) | methyltransferase type 11 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0575c | - | 2e-05 | 43.75% (48) | benzoquinone methyltransferase |
| M. gilvum PYR-GCK | Mflv_4471 | - | 7e-05 | 38.33% (60) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv0560c | - | 2e-05 | 43.75% (48) | benzoquinone methyltransferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4696c | - | 1e-05 | 33.58% (137) | methyltransferase |
| M. marinum M | MMAR_0460 | - | 1e-76 | 59.48% (232) | hypothetical protein MMAR_0460 |
| M. avium 104 | MAV_4317 | - | 1e-05 | 33.61% (122) | putative methyltransferase |
| M. smegmatis MC2 155 | MSMEG_0997 | - | 3e-84 | 70.39% (206) | hypothetical protein MSMEG_0997 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0662 | - | 2e-07 | 38.46% (78) | methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0888|M.vanbaalenii_PYR-1 ---------------------MFGQLYDRALDGERCWVRHDDGRVKNLPV
MSMEG_0997|M.smegmatis_MC2_155 ---------------------MHGHLYDRALDGERCWIRHADGRVQRLPV
MMAR_0460|M.marinum_M -------MDVRPGRARCKGREMLGHLYERALGGERCWIRHEDGELRPLPT
Mb0575c|M.bovis_AF2122/97 ---------MSTVLTYIRAVDIYEHMTESLDLEFESAYRGESVAFGEGVR
Rv0560c|M.tuberculosis_H37Rv ---------MSTVLTYIRAVDIYEHMTESLDLEFESAYRGESVAFGEGVR
MUL_0662|M.ulcerans_Agy99 -------------------------MTESLDLQFATAYRGESAEFGAGIR
Mflv_4471|M.gilvum_PYR-GCK ----------------------MDMDQTPTLSRFDDFYKDEDK------T
MAB_4696c|M.abscessus_ATCC_199 MMFGTREVFEYFSCSACDTLQIVNVLEGEDLMRHYPATYYSHNGSGQPSA
MAV_4317|M.avium_104 ---------------------MTRSRHDRSLSFGSAAAAYERGRPS----
Mvan_0888|M.vanbaalenii_PYR-1 RRWLGG------------------------------SGGDATFDHAVVGL
MSMEG_0997|M.smegmatis_MC2_155 RRWLGG------------------------------ARSERRFDRTVVNL
MMAR_0460|M.marinum_M HRWLGAGAAERGCASAEGSSSGDVGTGDED------AASDEDFDEAVAQM
Mb0575c|M.bovis_AF2122/97 PPWSIG------------------------------EPQPELAALIVQGK
Rv0560c|M.tuberculosis_H37Rv PPWSIG------------------------------EPQPELAALIVQGK
MUL_0662|M.ulcerans_Agy99 PPWSLG------------------------------EPQPELATLIEQGK
Mflv_4471|M.gilvum_PYR-GCK PPWVIG------------------------------EPQPAVVDIERAGL
MAB_4696c|M.abscessus_ATCC_199 LQWLVTQRDRRKLDGGGRVFGALMSRPIPEGIFRVLLGGDAVNMLAELGI
MAV_4317|M.avium_104 ------------------------------------YPPEAIDWLLPVGA
Mvan_0888|M.vanbaalenii_PYR-1 -CHGPTIDLGCGPGRLVAHLVQRGVPALGVDLSATAVELARRSGAP----
MSMEG_0997|M.smegmatis_MC2_155 -CSGPTIDLGCGPGRLVAGLVRRGVPALGVDQSVAAVAIARNSGVP----
MMAR_0460|M.marinum_M -CSGPTIELGCGPGRLVARLVQRGIPALGIDRSVTAIRLAGRGGAP----
Mb0575c|M.bovis_AF2122/97 -FRGDVLDVGCGEAAISLALAERGHTTVGLDLSPAAVELARHEAAKRGLA
Rv0560c|M.tuberculosis_H37Rv -FRGDVLDVGCGEAAISLALAERGHTTVGLDLSPAAVELARHEAAKRGLA
MUL_0662|M.ulcerans_Agy99 -VHGEVLDAGCGEAALAMALAGRGHPVVGLDISPTAVELAGREAARPGLT
Mflv_4471|M.gilvum_PYR-GCK -ITGRVLDVGCGTGERTILLTAAGYDVLGVDGAPSAVEQARHNAKTR-VV
MAB_4696c|M.abscessus_ATCC_199 GRDARILDVGCGSGALLDRLARVGFSSLLGADPFIAADGQSREGIP----
MAV_4317|M.avium_104 ---RQVLDLGAGTGKLTTRLVERGLDVVAVDPIPDMLEVLRSSLPETRAL
:: *.* . *. * :
Mvan_0888|M.vanbaalenii_PYR-1 -------------------------ALRRDVFEPLPGTGRWQTVLLADGN
MSMEG_0997|M.smegmatis_MC2_155 -------------------------VLCRDLFGPLPGTGRWHTALLADGN
MMAR_0460|M.marinum_M -------------------------AILGDVFEPLPGMGRWQTVLLVDGN
Mb0575c|M.bovis_AF2122/97 NASFEVADASSFTGY----DGRFDTIVDSTLFHSMPVESREGYLQSIVRA
Rv0560c|M.tuberculosis_H37Rv NASFEVADASSFTGY----DGRFDTIVDSTLFHSMPVESREGYLQSIVRA
MUL_0662|M.ulcerans_Agy99 NASFAVADITDFASYPPESAGRFNTIMDSTLFHSMPVELREGCQRSIVRA
Mflv_4471|M.gilvum_PYR-GCK DARFDVADALDLGDE-----PAYDTIIDSALFHIFDDADREKYVRSLGRA
MAB_4696c|M.abscessus_ATCC_199 --------------------------LLRRYLGEVTG---EFDLVMFNHS
MAV_4317|M.avium_104 LGTAEEIPLED---------NSVDVVLVAQAWHWVDPERAIPEVARVLRP
: .
Mvan_0888|M.vanbaalenii_PYR-1 VGLGGDPRRVLGRATELLRRGGRCLAELDGTTSGVDVG----WVRLES-H
MSMEG_0997|M.smegmatis_MC2_155 VGLGGDPWRVLQRAGELLRRGGECLAEFDTDITGVEAN----WVRLES-A
MMAR_0460|M.marinum_M VGLGGDPLRILGRAAELLSRGGRCVVEFDAQTIGVCSR----WVRLES-A
Mb0575c|M.bovis_AF2122/97 AAPGASYFVLVFDRAAIP-EGPINAVTEDELRAAVSKY----WIIDEIKP
Rv0560c|M.tuberculosis_H37Rv AAPGASYFVLVFDRAAIP-EGPINAVTEDELRAAVSKY----WIIDEIKP
MUL_0662|M.ulcerans_Agy99 AAPGASYFVLVFDKAALSGDGPINGVTEEELRDAVSKH----WVIDEIRP
Mflv_4471|M.gilvum_PYR-GCK TAPGSVVHLLALSD---TGRGFGPEVSEETIRAAFADPG---WDIEALTT
MAB_4696c|M.abscessus_ATCC_199 LEHMPDPVSTLQIAARKLTTGGMCLARVPTTSSEAWSVYGADWVQADAPR
MAV_4317|M.avium_104 GGRLGLVWNTRDERLGWVRELGRIIGSDGDGRTHVALPEP--FTDLAHHE
:
Mvan_0888|M.vanbaalenii_PYR-1 RMIGPWFRWASVGID--------------------CVPSLAAEAGLAVGG
MSMEG_0997|M.smegmatis_MC2_155 HAVGPWFRWASVGLD--------------------CAEHIAADVGLALST
MMAR_0460|M.marinum_M RDVGPWFRWASVGVD--------------------SAAMLAAQVGLTLIG
Mb0575c|M.bovis_AF2122/97 ARLYARFPAGFAGMP----------------------ALLDIR--EEPNG
Rv0560c|M.tuberculosis_H37Rv ARLYARFPAGFAGMP----------------------ALLDIR--EEPNG
MUL_0662|M.ulcerans_Agy99 ARIHANLPPEVPGCPPS------------TLATNPRAASPSVRGCSQPIW
Mflv_4471|M.gilvum_PYR-GCK TYYRGVITAPHADAMG----------------------LDVGTVVDEPAW
MAB_4696c|M.abscessus_ATCC_199 HMVIPSRRGMALAAERAGLRVLRAFDDSTFGQFTGSEAYRADVPVTDPKI
MAV_4317|M.avium_104 VEWTNYLTPQALIDLVAS-----------------RSYCITSPTEVRTRT
Mvan_0888|M.vanbaalenii_PYR-1 IHRIGTRVVADLVA---------------------
MSMEG_0997|M.smegmatis_MC2_155 VHSIGDRAVASLVAA--------------------
MMAR_0460|M.marinum_M VRMLGGRVIANLAAL--------------------
Mb0575c|M.bovis_AF2122/97 LQSIGGWLLS-AHLG--------------------
Rv0560c|M.tuberculosis_H37Rv LQSIGGWLLS-AHLG--------------------
MUL_0662|M.ulcerans_Agy99 ADGTPHAVYSPANEG--------------------
Mflv_4471|M.gilvum_PYR-GCK LARIRRL----------------------------
MAB_4696c|M.abscessus_ATCC_199 LTMFRPKQIWEWEKRAKRLNQQHRGDQTGFVLRVK
MAV_4317|M.avium_104 LDQVRHLLATHPALANSTGLALPYVTRCTRATLAG