For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDVRPGRARCKGREMLGHLYERALGGERCWIRHEDGELRPLPTHRWLGAGAAERGCASAEGSSSGDVGTG DEDAASDEDFDEAVAQMCSGPTIELGCGPGRLVARLVQRGIPALGIDRSVTAIRLAGRGGAPAILGDVFE PLPGMGRWQTVLLVDGNVGLGGDPLRILGRAAELLSRGGRCVVEFDAQTIGVCSRWVRLESARDVGPWFR WASVGVDSAAMLAAQVGLTLIGVRMLGGRVIANLAAL
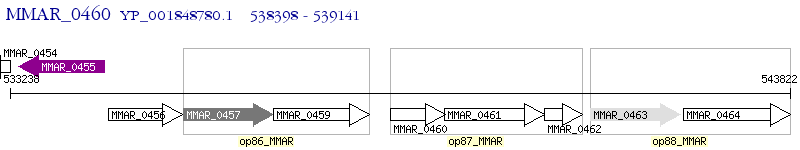
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0460 | - | - | 100% (247) | hypothetical protein MMAR_0460 |
| M. marinum M | MMAR_1164 | - | 6e-05 | 33.01% (103) | methyltransferase |
| M. marinum M | MMAR_0909 | - | 1e-04 | 45.24% (42) | methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0997 | - | 7e-72 | 56.09% (230) | hypothetical protein MSMEG_0997 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2851 | - | 4e-05 | 33.01% (103) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_0888 | - | 1e-76 | 59.48% (232) | methyltransferase type 12 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0997|M.smegmatis_MC2_155 --------------MHGHLYDRALDGERCWIRHADGRVQRLP-VRRWLGG
Mvan_0888|M.vanbaalenii_PYR-1 --------------MFGQLYDRALDGERCWVRHDDGRVKNLP-VRRWLGG
MMAR_0460|M.marinum_M MDVRPGRARCKGREMLGHLYERALGGERCWIRHEDGELRPLP-THRWLGA
MUL_2851|M.ulcerans_Agy99 -------------MVEQSIWMQKVAADPGHSRWYIERFRAMARAGEDLVG
: :: : : .: * ..: :. . . * .
MSMEG_0997|M.smegmatis_MC2_155 ------------------------ARSERRFDRTVVNLCSGPTIDLGCGP
Mvan_0888|M.vanbaalenii_PYR-1 ------------------------SGGDATFDHAVVGLCHGPTIDLGCGP
MMAR_0460|M.marinum_M GAAERGCASAEGSSSGDVGTGDEDAASDEDFDEAVAQMCSGPTIELGCGP
MUL_2851|M.ulcerans_Agy99 ---------------------------EARLVDSIAPRG-AHILDAGCGP
: : ::. . :: ****
MSMEG_0997|M.smegmatis_MC2_155 GRLVAGLVRRGVPALGVDQSVAAVAIARNS--GVPVLCRDLFG---PLPG
Mvan_0888|M.vanbaalenii_PYR-1 GRLVAHLVQRGVPALGVDLSATAVELARRS--GAPALRRDVFE---PLPG
MMAR_0460|M.marinum_M GRLVARLVQRGIPALGIDRSVTAIRLAGRG--GAPAILGDVFE---PLPG
MUL_2851|M.ulcerans_Agy99 GRLGRHLAAAGHRVVGVDVDPALIEAAEQDYPGPQWLVGDLAELDLPARG
*** *. * .:*:* . : : * .. * : *: * *
MSMEG_0997|M.smegmatis_MC2_155 TGRWHTALLADGNVGLGGDP---WRVLQRAGELLRRGGECLAEFDTDITG
Mvan_0888|M.vanbaalenii_PYR-1 TGRWQTVLLADGNVGLGGDP---RRVLGRATELLRRGGRCLAELDGTTSG
MMAR_0460|M.marinum_M MGRWQTVLLVDGNVGLGGDP---LRILGRAAELLSRGGRCVVEFDAQTIG
MUL_2851|M.ulcerans_Agy99 IAEPFDVIVSAGNVMTFLAPSTRARVLGRLRAHLCSDGRAVIGFGAGRDY
.. .:: *** * *:* * * .*..: :.
MSMEG_0997|M.smegmatis_MC2_155 VEANWVRLESAHAVGPWFRWASVGLDCAEHIAADVGLALSTVHSIGDRAV
Mvan_0888|M.vanbaalenii_PYR-1 VDVGWVRLESHRMIGPWFRWASVGIDCVPSLAAEAGLAVGGIHRIGTRVV
MMAR_0460|M.marinum_M VCSRWVRLESARDVGPWFRWASVGVDSAAMLAAQVGLTLIGVRMLGGRVI
MUL_2851|M.ulcerans_Agy99 AFADFLDDAQAAGLVPDLLLSTWDLR---PFTDDCDFLVALLRPA-----
. :: . : * : :: .: :: : .: : ::
MSMEG_0997|M.smegmatis_MC2_155 ASLVAA
Mvan_0888|M.vanbaalenii_PYR-1 ADLVA-
MMAR_0460|M.marinum_M ANLAAL
MUL_2851|M.ulcerans_Agy99 ------