For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTTQMRRGEIDAVVDKAAAAAAEFRKLDQAQVDAIVEAMVRAGIRAAAALAQVAIEETGFGVFEDKVVK NYVATEFLHDYLRDKKSVGVVEEDIENNIVRVAEPIGVVLAITPVTNPTSTVLFKAIVAAKTRNAIVFRP SPYAVRCCERSVEILREAAEAAGMPTGALQVIPDEAHEVTHYLFAHEKIDFLWVTGGPKIVALANAAGKP GLCVGPGNAPIYIHKTADVRGAVVDILISKTFDSSVICPAEQTCVIDDEIYDAMIAEFERMGARLLTEEE TSAIARFAFGCGDKIALEALGQKAPELAARAGFSVPPTVKVLLAALPADLEELAAHPLVQEKLMPVLGVV RARSVDHAIDVAVLVTEHGGLGHTSAVYANDQAVVDAYSAAVRTGRILVNAPTAVGALGGVYNNLTPTFS LGCGTWGGSRTTENVNYRQLLNIKTVAQRRTPPQWFRVPSNTYFNAGALENLRDLDCETVVIVTDAASDE RGVVDLLRGKLRTRHVQVFSEVPPEPDESMIRRGVQLLERVQPEVLIAVGGGSVLDAGKAMRLFFEHPEK TLDELTMPFLDPRKRVADYPTDRHRLQLVAVPTTSGTGSEVSPAAVLTVRGRKQTLVDYSLVPDLAIVDP VLTSSMPPVLTADTGIDALTHALEAVVSIFASPYTDALCAQAARLIFDALPRAYRDPEDLAARTDMSNAA TLAGLAFSNAFVGTNHALAHAVGAKFGIAHGRANAIFLPHVLRYNASLPSKFMPAPGYSAYIAPDKYAQI GQLIFGGHEPEDSRGRLFLGVQDLLEQLDMPGSLKAIGVSEDAFTAALPELAMTAFEDLSNRTNPRMPLV SEISDLLRLGYYGSEAPG
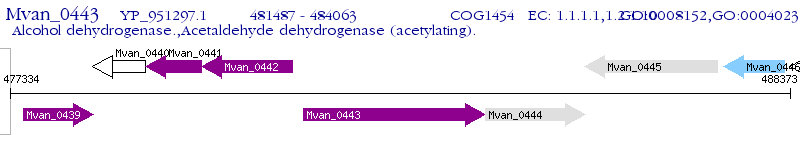
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0443 | - | - | 100% (858) | bifunctional acetaldehyde-CoA/alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5566 | - | 1e-71 | 36.64% (434) | aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5476 | - | 1e-45 | 32.98% (382) | iron-containing alcohol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0775c | mmsA | 8e-10 | 25.00% (348) | methylmalonate-semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1242 | - | 4e-70 | 35.71% (434) | aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0753c | mmsA | 1e-10 | 25.29% (348) | methylmalonate-semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_01458 | proA | 4e-12 | 26.16% (279) | gamma-glutamyl phosphate reductase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 2e-12 | 25.84% (267) | gamma-aminobutyraldehyde dehydrogenase |
| M. marinum M | MMAR_4110 | - | 0.0 | 85.11% (853) | aldehyde dehydrogenase |
| M. avium 104 | MAV_1747 | proA | 4e-13 | 25.45% (279) | gamma-glutamyl phosphate reductase |
| M. smegmatis MC2 155 | MSMEG_0276 | - | 9e-72 | 36.20% (453) | aldehyde-alcohol dehydrogenase |
| M. thermoresistible (build 8) | TH_3419 | - | 1e-13 | 25.56% (446) | PUTATIVE putative aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_3976 | - | 0.0 | 84.53% (853) | bifunctional acetaldehyde-CoA/alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3328|M.abscessus_ATCC_1997 --------MSVPSSWINGR-PVATHGDTHRVINPATGEAVADLKLATTAD
TH_3419|M.thermoresistible__bu VQRTLPDKLPAPRLVIGGGDVISTSGGVYEHHYAATGGVQAEVPLAGPAE
Mb0775c|M.bovis_AF2122/97 -------MTTQISHFIDGQRTAGQSTRSADVFDPNTGQIQAKVPMAGKSD
Rv0753c|M.tuberculosis_H37Rv -------MTTQISHFIDGQRTAGQSTRSADVFDPNTGQIQAKVPMAGKSD
MMAR_4110|M.marinum_M --------------------------MTTTPTESASPVEAPAIDNLRTAE
MUL_3976|M.ulcerans_Agy99 --------------------------MTTTPTESASPVEAPAIDNLRTAE
Mvan_0443|M.vanbaalenii_PYR-1 --------------------------MSTT--------------QMRRGE
Mflv_1242|M.gilvum_PYR-GCK --------------------------------------------MIAVPH
MSMEG_0276|M.smegmatis_MC2_155 ----------------------------------MSEVIQDRRSAVDTAR
MLBr_01458|M.leprae_Br4923 ----------------------------------------------MRQE
MAV_1747|M.avium_104 --------------------MGLQAPFRSGGGTSRPQGRAAPRAVDLRRE
MAB_3328|M.abscessus_ATCC_1997 VDAAVAAARAAGPAWASSTPVDRSSVLAKLAAVLSANADTLVADEVAQTG
TH_3419|M.thermoresistible__bu VDAAVAAARAAFPAWRDLDLNRRRDILQELARRLVADGERLGAIATLEVG
Mb0775c|M.bovis_AF2122/97 IDAAVASAVEAQKGWAAWNPQRRARVLMRFIELVNDTIDELAELLSREHG
Rv0753c|M.tuberculosis_H37Rv IDAAVASAVEAQKGWAAWNPQRRARVLMRFIELVNDTIDELAELLSREHG
MMAR_4110|M.marinum_M IDAIVDTAAEAARAFRKLDQQQVDAIVEAMVRAGVRAAGELAGVAIEETG
MUL_3976|M.ulcerans_Agy99 IDAIVDTAAEAARAFRKLDQQQVDAIVEAMVRAGVRAAGELAGVAIEETG
Mvan_0443|M.vanbaalenii_PYR-1 IDAVVDKAAAAAAEFRKLDQAQVDAIVEAMVRAGIRAAAALAQVAIEETG
Mflv_1242|M.gilvum_PYR-GCK AGQLLERAHFAASAYADYDQASVARIVDAVAEAGHRNAERFAAAAVAETQ
MSMEG_0276|M.smegmatis_MC2_155 AGHMLERARWAAAAYGEYDAATVEGIVAAVAEAAHAHAERFAADAVAETG
MLBr_01458|M.leprae_Br4923 VHDAARRARVAARALAVMPTVAKDHALRAAATAVTAHTEEILAANAEDLQ
MAV_1747|M.avium_104 VHDAARRARVASRVLALLPTVAKDQALRSAADAIAGHAELILSANAEDLE
* * : : :
MAB_3328|M.abscessus_ATCC_1997 KP-VRLASEFDVPGSIDNVEFFAGAARHLEGKATAEYSADHTSSIRREAA
TH_3419|M.thermoresistible__bu TP-NLLASS-SSAMAADWFTYYAGWIGRSAGEVVPVPAGGALDYVLDEPL
Mb0775c|M.bovis_AF2122/97 KT-LADARG-DVQRGIEVIEFCLGIPHLLKGEYTEGAGPGIDVYSLRQPL
Rv0753c|M.tuberculosis_H37Rv KT-LADARG-DVQRGIEVIEFCLGIPHLLKGEYTEGAGPGIDVYSLRQPL
MMAR_4110|M.marinum_M FG----VFEDKVVKNYVATEFLHDYLRGKKSVGVIDEDVEHNIVYVAEPI
MUL_3976|M.ulcerans_Agy99 FG----VFEDKVVKNYVATEFLHDYLRGKKSVGVIDEDVEHNIVYVAEPI
Mvan_0443|M.vanbaalenii_PYR-1 FG----VFEDKVVKNYVATEFLHDYLRDKKSVGVVEEDIENNIVRVAEPI
Mflv_1242|M.gilvum_PYR-GCK MG----VVEHKVTKNRACSRGIVEFYRDQ-DYVTPRIDTARKIVEIPRPA
MSMEG_0276|M.smegmatis_MC2_155 MG----VVADKVRKNRACSRGIVEYYRGH-DYISPRVDPDRKIVELPRPA
MLBr_01458|M.leprae_Br4923 AARATDTLAATLDRLALDRQRIDQIAGGLRQVAALPDPIGEVLRGYTLPN
MAV_1747|M.avium_104 TARAAGTPSAMLDRLALDTKRVEGIAAGLRQVAGLPDPVGEVLRGYTLPN
. .
MAB_3328|M.abscessus_ATCC_1997 GVVATITPWNYPLQMAVWK---------VLPALAAGCTVVIKPSELTP--
TH_3419|M.thermoresistible__bu GVIAAIIPWNGPLISIGMK---------VAPALAAGNCVVLKPPENAP--
Mb0775c|M.bovis_AF2122/97 GVVAGITPFNFPAMIPLWK---------AGPALACGNAFVLKPSERDP--
Rv0753c|M.tuberculosis_H37Rv GVVAGITPFNFPAMIPLWK---------AGPALACGNAFVLKPSERDP--
MMAR_4110|M.marinum_M GVVLGITPVTNPTSTVLFK---------AIVAAKTRNAILFRPSPYAVRC
MUL_3976|M.ulcerans_Agy99 GVVLGITPVTNPTSTVLFK---------AIVAAKTRNAILFRPSPYAVRC
Mvan_0443|M.vanbaalenii_PYR-1 GVVLAITPVTNPTSTVLFK---------AIVAAKTRNAIVFRPSPYAVRC
Mflv_1242|M.gilvum_PYR-GCK GVVLALTPTTNPVATVYFK---------VILALMTRNAVVVAPHPRARQC
MSMEG_0276|M.smegmatis_MC2_155 GVVLALTPTTNPVATVYFK---------TILALMTRNAVVVAPHPRAARC
MLBr_01458|M.leprae_Br4923 GLQLRQQRVPLGVVGMIYEGRPNITVDAFGLAFKSGNAALLRGSSSAAKS
MAV_1747|M.avium_104 GLQLRQQRVPLGVVGMIYEGRPNVTVDAFGLALKSGNAVLLRGSSSATRS
*: : * :.
MAB_3328|M.abscessus_ATCC_1997 --LTTLTLARLAAEAGLPEGVLNVVTGRGDDVGYALASHP-DVDLVTFTG
TH_3419|M.thermoresistible__bu --FSALRFAELAAEAGLPPGVLNVIPG-GAEAGSALCSHP-GVDKITFTG
Mb0775c|M.bovis_AF2122/97 --SVPVRLAELFIEAGLPAGVFQVVHG-DKEAVDAILHHP-DIKAVGFVG
Rv0753c|M.tuberculosis_H37Rv --SVPVRLAELFIEAGLPAGVFQVVHG-DKEAVDAILHHP-DIKAVGFVG
MMAR_4110|M.marinum_M CEQSIEILRTAAEAAGMPPGALQVIPDAAHEVTHYLFKHP-KVDFIWVTG
MUL_3976|M.ulcerans_Agy99 CEQSIEILRTAAEAAGMPPGALQVIPDAAHEVTHYLFKHP-KVDFIWVTG
Mvan_0443|M.vanbaalenii_PYR-1 CERSVEILREAAEAAGMPTGALQVIPDEAHEVTHYLFAHE-KIDFLWVTG
Mflv_1242|M.gilvum_PYR-GCK SADAARVLAEAAEAAGAPDGIVQVIDEPSLPLVEALMSDE-RTDVIVATG
MSMEG_0276|M.smegmatis_MC2_155 SAEAARVLAEAAVAAGAPDGVVQVIDEPSIPLVQALMADE-RTDVIVATG
MLBr_01458|M.leprae_Br4923 NQALVEVLHAALVSEDLPADAVQLLSAADRSTVTHLIQARGLVDVVIPRG
MAV_1747|M.avium_104 NQALVQVLRASLVSEDLPADAVQLLSADDRATVTHLIQARGLVDVVIPRG
: . * . .::: : . : *
MAB_3328|M.abscessus_ATCC_1997 STAVGRKVMAAAAVHGHRTQLELGGKAPFVVFDDADMDAAIAGAV-AGAI
TH_3419|M.thermoresistible__bu GGETARKVFAAAAQALTPVVLELGGKSANIVFPDADLDAAATMAVSAGLM
Mb0775c|M.bovis_AF2122/97 SSDIAQYIYAGAAATGKRAQCFGGAKNHMIVMPDADLDQAVDALIGAGYG
Rv0753c|M.tuberculosis_H37Rv SSDIAQYIYAGAAATGKRAQCFGGAKNHMIVMPDADLDQAVDALIGAGYG
MMAR_4110|M.marinum_M G---PKIVALAN-SAGKPGLSVGPGNAPIYIHKTADLKGAVVDILISKTF
MUL_3976|M.ulcerans_Agy99 G---PKIVALAN-SAGKPGLSVGPGNAPIYIHKTADLKGAVVDILISKTF
Mvan_0443|M.vanbaalenii_PYR-1 G---PKIVALAN-AAGKPGLCVGPGNAPIYIHKTADVRGAVVDILISKTF
Mflv_1242|M.gilvum_PYR-GCK G---TGVVRAAY-SSGNPALGVGPGNVPVFVDATADIDAAARRIVDSKAF
MSMEG_0276|M.smegmatis_MC2_155 G---TAVVRAAY-SSGNPALGVGPGNVPVLVDASADVAAAAQRIVDSKAF
MLBr_01458|M.leprae_Br4923 G---ANLIDAVVRDAQVPIIETGVGNCHVYVHQAADLEVAERILLNSKTR
MAV_1747|M.avium_104 G---ANLINAVVRDAQVPTIETGVGNCHVYVHEGADLEVAERVLLNSKTR
. : .: : **: * : :
MAB_3328|M.abscessus_ATCC_1997 INSGQDCTAATRALVARDLYDDFVAGVGEVMSKVVVGDPLDPDTDIGPLI
TH_3419|M.thermoresistible__bu MLSGQGCILPTRLLVHDDVYDEMVTRVVAGADALTVGDPWQPTTMMGPLA
Mb0775c|M.bovis_AF2122/97 SAGERCMAISVAVPVGDQTAERLRARLIERINNLRVGHSLDPKADYGPLV
Rv0753c|M.tuberculosis_H37Rv SAGERCMAISVAVPVGDQTAERLRARLIERINNLRVGHSLDPKADYGPLV
MMAR_4110|M.marinum_M DS-SVICPAEQTCIIDDEIYDEMIAEFERMGAQLLTADQAKAVAEFAFGC
MUL_3976|M.ulcerans_Agy99 DS-SVICPAKQTCIIDDEIYDEMIAEFERMGAQLLTADQAKAVAEFAFGC
Mvan_0443|M.vanbaalenii_PYR-1 DS-SVICPAEQTCVIDDEIYDAMIAEFERMGARLLTEEETSAIARFAFGC
Mflv_1242|M.gilvum_PYR-GCK DN-SVLCTNESVLIAEESIAAKLRTSLTRHGAHILDADAADRLREYMFPD
MSMEG_0276|M.smegmatis_MC2_155 DN-SVLCTNESVLIAEDAVADKLRAALTRAGAHILDEDGARRLRAYMFAD
MLBr_01458|M.leprae_Br4923 R--PSVCNAAETLLIDAAIADHTMPRLVAA--------------------
MAV_1747|M.avium_104 R--PSVCNAAETLLVDAAIAEEALPRLVTA--------------------
. .
MAB_3328|M.abscessus_ATCC_1997 SAAHRAKVSSIVDRAP-AQGGRIVTGATAPD--------------LPGSF
TH_3419|M.thermoresistible__bu TAAQLERVTRAVDSALSDKAGKLLAGGRRPDG------------LDAGYF
Mb0775c|M.bovis_AF2122/97 TGAALARVRDYIGQGVAAGAELVVDGRDRASDDLTFGLPEGDANLEGGFF
Rv0753c|M.tuberculosis_H37Rv TGAALARVRDYIGQGVAAGAELVVDGRDRASDDLTFGLPEGDANLEGGFF
MMAR_4110|M.marinum_M GDKVSLDAVGQKAPELAARAGFSVPPTVKVLL---------------AQL
MUL_3976|M.ulcerans_Agy99 GDKVSLDAVGQKAPELAARAGFSVPPTVKVPL---------------AQL
Mvan_0443|M.vanbaalenii_PYR-1 GDKIALEALGQKAPELAARAGFSVPPTVKVLL---------------AAL
Mflv_1242|M.gilvum_PYR-GCK G-ALNTEVVGRDAAWIAERIGLRVTPKTRVLI---------------APF
MSMEG_0276|M.smegmatis_MC2_155 G-HLNTDVVGRDAAWIAGQAGLRVTPKTRVLI---------------APF
MLBr_01458|M.leprae_Br4923 ----------LQGAGVTVHGGLSGDPDK----------------------
MAV_1747|M.avium_104 ----------LQDAGVTVHGGFARDATE----------------------
MAB_3328|M.abscessus_ATCC_1997 YRPTLIADVAETSEVYRDEIFGPVLTVRPFTDDDDALRQANDTAYGLAAS
TH_3419|M.thermoresistible__bu YRPTVFGDVDPASDLAQTEIFGPVLAILRFRSEEEAVELANNTQYGLAAY
Mb0775c|M.bovis_AF2122/97 IGPTLFDHVAAHMSIYTDEIFGPVLCMVRARDYEEALRLPSEHEYGNGVA
Rv0753c|M.tuberculosis_H37Rv IGPTLFDHVAAHMSIYTDEIFGPVLCMVRARDYEEALRLPSEHEYGNGVA
MMAR_4110|M.marinum_M PSDLGELAAHPLVQEKLMPVLGVVRARSVQHGIDAAVLVTEHGGLGHTSA
MUL_3976|M.ulcerans_Agy99 PSDLDELAAHPLVQEKLMPVLGVVRARSVQHGIDAAVLVTEHGGLGHTSA
Mvan_0443|M.vanbaalenii_PYR-1 PADLEELAAHPLVQEKLMPVLGVVRARSVDHAIDVAVLVTEHGGLGHTSA
Mflv_1242|M.gilvum_PYR-GCK T---DVIGEEVLTHEKLCPVLGMTTVADAPRGIRAARAVVRIAGAGHSAA
MSMEG_0276|M.smegmatis_MC2_155 D---HVIAEEMLAHEKLSPVLGMTTVADAARGIRAARAVVRIGGAGHSAA
MLBr_01458|M.leprae_Br4923 -------------ADLCREYLSMDIAVAVVDGVDAAIAHINEYGTGHTEA
MAV_1747|M.avium_104 -------------DDLRREYLSMDIAVAVVDGVDAAIAHINEYGTGHTEA
:. * *
MAB_3328|M.abscessus_ATCC_1997 AWTRDVYRAQRASREIRAGCVWINDHIPIISEMP-HGGFGASGFG--KDM
TH_3419|M.thermoresistible__bu IHTRDLRRAHVMAAALDAGGVAVNGFPIVPSAAP-FGGVKQSGFG--REG
Mb0775c|M.bovis_AF2122/97 IFTRDGDAARDFVSRGQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQH
Rv0753c|M.tuberculosis_H37Rv IFTRDGDAARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQH
MMAR_4110|M.marinum_M IYANDDDVIEAYGLAVRTGRILVNAPTAVGALGGVYNNLTPTFSLGCGTW
MUL_3976|M.ulcerans_Agy99 IYANDDDVIEAYGLAVRTGRILVNAPTAVGALGGVYNNLTPTFSLGCGTW
Mvan_0443|M.vanbaalenii_PYR-1 VYANDQAVVDAYSAAVRTGRILVNAPTAVGALGGVYNNLTPTFSLGCGTW
Mflv_1242|M.gilvum_PYR-GCK IHSEDASVITEFAAKVPVLRVSVNVGNSTGSSG-LDTNLAPSMTIGTGFV
MSMEG_0276|M.smegmatis_MC2_155 IHSENPAVITDFAAQVPVLRVSVNVGNSTGSAG-LDTNLAPSMTIGTGFV
MLBr_01458|M.leprae_Br4923 IVTTNMAAAQRFADGVDSATVMVNASTAFTDGGQFGFGAEIGISTQKLHA
MAV_1747|M.avium_104 IVTTNMAAAQRFTDGVDAAAVMVNASTAFTDGEQFGFGAEIGISTQKLHA
: : : :* .
MAB_3328|M.abscessus_ATCC_1997 SDYSLEEYLTVKHVMSDITGTADKDWHRTIFAKR----------------
TH_3419|M.thermoresistible__bu GIWGLREFQRTKNVYIGLQ-------------------------------
Mb0775c|M.bovis_AF2122/97 GPAAIQFYTKVKTVTSRWPSGIKDGAEFVIPTMS----------------
Rv0753c|M.tuberculosis_H37Rv GPAAIQFYTKVKTVTSRWPSGIKDGAEFVIPTMS----------------
MMAR_4110|M.marinum_M GGSSTTENVNYRQLLNIKTVSRRRTPPQWFRVPSNTYFNEGALDNLRELD
MUL_3976|M.ulcerans_Agy99 GGSSTTENVNYRQLLNIKTVSRRRTPPQWFRVPSNTYFNEGALDNLRELD
Mvan_0443|M.vanbaalenii_PYR-1 GGSRTTENVNYRQLLNIKTVAQRRTPPQWFRVPSNTYFNAGALENLRDLD
Mflv_1242|M.gilvum_PYR-GCK GRSSIGENLQPENLLNWARIAYNADTAVTMPNFAG---------------
MSMEG_0276|M.smegmatis_MC2_155 GRSSIGENLQPHNLVNWTRIAYNSDPGVVMGNFTG---------------
MLBr_01458|M.leprae_Br4923 RGPMGLSEMTSTKWIVWGDGQIRPA-------------------------
MAV_1747|M.avium_104 RGPMGLPELTSTKWIAWGDGQTRPA-------------------------
MAB_3328|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3419|M.thermoresistible__bu --------------------------------------------------
Mb0775c|M.bovis_AF2122/97 --------------------------------------------------
Rv0753c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4110|M.marinum_M CETVVVVTDALTEERGVVEALRGKLRAQHVQVFSEVTPEPDETTIRRGVA
MUL_3976|M.ulcerans_Agy99 CETVVVVTDALTEERGVVEALRGKLRAQHVQVFSEVTPEPDETTIRRGVA
Mvan_0443|M.vanbaalenii_PYR-1 CETVVIVTDAASDERGVVDLLRGKLRTRHVQVFSEVPPEPDESMIRRGVQ
Mflv_1242|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0276|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01458|M.leprae_Br4923 --------------------------------------------------
MAV_1747|M.avium_104 --------------------------------------------------
MAB_3328|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3419|M.thermoresistible__bu --------------------------------------------------
Mb0775c|M.bovis_AF2122/97 --------------------------------------------------
Rv0753c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4110|M.marinum_M LMQRVQPDLLIAIGGGSVLDAGKAMRLFYEHPEKSLDALTMPFLDPRKRV
MUL_3976|M.ulcerans_Agy99 LMQRVQPDLLIAIGGGSVLDAGKAMRLFYEHPEKSLDALTMPFLDPRKRV
Mvan_0443|M.vanbaalenii_PYR-1 LLERVQPEVLIAVGGGSVLDAGKAMRLFFEHPEKTLDELTMPFLDPRKRV
Mflv_1242|M.gilvum_PYR-GCK ---------------------------------------LSPWHAPTGPV
MSMEG_0276|M.smegmatis_MC2_155 ---------------------------------------IDPWRAPAGPV
MLBr_01458|M.leprae_Br4923 --------------------------------------------------
MAV_1747|M.avium_104 --------------------------------------------------
MAB_3328|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3419|M.thermoresistible__bu --------------------------------------------------
Mb0775c|M.bovis_AF2122/97 --------------------------------------------------
Rv0753c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4110|M.marinum_M ADYPTDRHRIQLVAVPTTSGTGSEVSPAAVLTVGGKKETLVDYSLVPDLA
MUL_3976|M.ulcerans_Agy99 ADYPTDRHRIQLVAVPTTSGTGSEVSPAAVLTVGGKKETLVDYSLVPDLA
Mvan_0443|M.vanbaalenii_PYR-1 ADYPTDRHRLQLVAVPTTSGTGSEVSPAAVLTVRGRKQTLVDYSLVPDLA
Mflv_1242|M.gilvum_PYR-GCK PAYPRASNDQS-ATPAPAPAAS----------------------------
MSMEG_0276|M.smegmatis_MC2_155 PAYPRASNDRDGAVIQAAPQRS----------------------------
MLBr_01458|M.leprae_Br4923 --------------------------------------------------
MAV_1747|M.avium_104 --------------------------------------------------
MAB_3328|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3419|M.thermoresistible__bu --------------------------------------------------
Mb0775c|M.bovis_AF2122/97 --------------------------------------------------
Rv0753c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4110|M.marinum_M IVDPVLTSSMPQKLTADTGIDALTHALEAIVSIFASPYTDALCAQAVRLI
MUL_3976|M.ulcerans_Agy99 IVDPVLTSSMPQKLTADIGIDALTHALEAIVSIFASPYTDALCAQAVRLI
Mvan_0443|M.vanbaalenii_PYR-1 IVDPVLTSSMPPVLTADTGIDALTHALEAVVSIFASPYTDALCAQAARLI
Mflv_1242|M.gilvum_PYR-GCK ------VSRPTAPRSADPGIEALRAELRALVAEELAQLIKR---------
MSMEG_0276|M.smegmatis_MC2_155 ------YQSP--SRTTDVNLDALRAELRALVVEELAQLIKR---------
MLBr_01458|M.leprae_Br4923 --------------------------------------------------
MAV_1747|M.avium_104 --------------------------------------------------
MAB_3328|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3419|M.thermoresistible__bu --------------------------------------------------
Mb0775c|M.bovis_AF2122/97 --------------------------------------------------
Rv0753c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4110|M.marinum_M FDALPRAYDDPDDLSARTSMSNAATLAGLAFSNAFVGTNHALAHAVGAKF
MUL_3976|M.ulcerans_Agy99 FDALPRAYDDPDDLSARTSMSNAATLAGLAFSNAFVGTNHTLAHAVGAKF
Mvan_0443|M.vanbaalenii_PYR-1 FDALPRAYRDPEDLAARTDMSNAATLAGLAFSNAFVGTNHALAHAVGAKF
Mflv_1242|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0276|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01458|M.leprae_Br4923 --------------------------------------------------
MAV_1747|M.avium_104 --------------------------------------------------
MAB_3328|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3419|M.thermoresistible__bu --------------------------------------------------
Mb0775c|M.bovis_AF2122/97 --------------------------------------------------
Rv0753c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4110|M.marinum_M SVPHGRANGIFLPHVLRYNASLPTKFMPAPGYSAYIAPDKYAQLGQLVFG
MUL_3976|M.ulcerans_Agy99 SVPHGRANGIFLPHVLRYNASLPTKFMPAPGYSAYIAPDKYAQLGQLVFG
Mvan_0443|M.vanbaalenii_PYR-1 GIAHGRANAIFLPHVLRYNASLPSKFMPAPGYSAYIAPDKYAQIGQLIFG
Mflv_1242|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0276|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01458|M.leprae_Br4923 --------------------------------------------------
MAV_1747|M.avium_104 --------------------------------------------------
MAB_3328|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3419|M.thermoresistible__bu --------------------------------------------------
Mb0775c|M.bovis_AF2122/97 --------------------------------------------------
Rv0753c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4110|M.marinum_M GHLPDDSRQRLFAGVDDLLDRLDMPRSLREFGVDEQEFLAALPTLAMTAF
MUL_3976|M.ulcerans_Agy99 GHLPDDSRQRLFAGVDDLLDRLDMPRSLREFGVDEQGFLVALPTLAMTAF
Mvan_0443|M.vanbaalenii_PYR-1 GHEPEDSRGRLFLGVQDLLEQLDMPGSLKAIGVSEDAFTAALPELAMTAF
Mflv_1242|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0276|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01458|M.leprae_Br4923 --------------------------------------------------
MAV_1747|M.avium_104 --------------------------------------------------
MAB_3328|M.abscessus_ATCC_1997 -------------------------------------------------
TH_3419|M.thermoresistible__bu -------------------------------------------------
Mb0775c|M.bovis_AF2122/97 -------------------------------------------------
Rv0753c|M.tuberculosis_H37Rv -------------------------------------------------
MMAR_4110|M.marinum_M EDLSNRTNPRMPLVSEITDLLRLGYYGDTGVGEPALAPIKPGDVDGRSV
MUL_3976|M.ulcerans_Agy99 EDLSNRTNPRMPLVSEITDLLRLGYYGDTGVGEPALAPIKPGDVDGRSV
Mvan_0443|M.vanbaalenii_PYR-1 EDLSNRTNPRMPLVSEISDLLRLGYYGSEAPG-----------------
Mflv_1242|M.gilvum_PYR-GCK -------------------------------------------------
MSMEG_0276|M.smegmatis_MC2_155 -------------------------------------------------
MLBr_01458|M.leprae_Br4923 -------------------------------------------------
MAV_1747|M.avium_104 -------------------------------------------------