For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGLQAPFRSGGGTSRPQGRAAPRAVDLRREVHDAARRARVASRVLALLPTVAKDQALRSAADAIAGHAEL ILSANAEDLETARAAGTPSAMLDRLALDTKRVEGIAAGLRQVAGLPDPVGEVLRGYTLPNGLQLRQQRVP LGVVGMIYEGRPNVTVDAFGLALKSGNAVLLRGSSSATRSNQALVQVLRASLVSEDLPADAVQLLSADDR ATVTHLIQARGLVDVVIPRGGANLINAVVRDAQVPTIETGVGNCHVYVHEGADLEVAERVLLNSKTRRPS VCNAAETLLVDAAIAEEALPRLVTALQDAGVTVHGGFARDATEDDLRREYLSMDIAVAVVDGVDAAIAHI NEYGTGHTEAIVTTNMAAAQRFTDGVDAAAVMVNASTAFTDGEQFGFGAEIGISTQKLHARGPMGLPELT STKWIAWGDGQTRPA
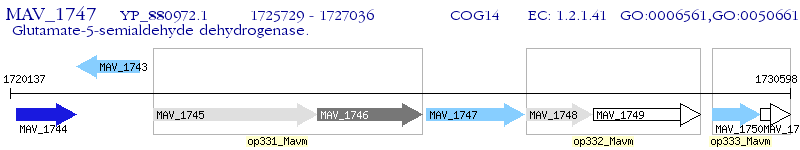
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1747 | proA | - | 100% (435) | gamma-glutamyl phosphate reductase |
| M. avium 104 | MAV_4265 | - | 8e-07 | 24.57% (468) | aldehyde dehydrogenase (NAD) family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2451c | proA | 0.0 | 83.37% (415) | gamma-glutamyl phosphate reductase |
| M. gilvum PYR-GCK | Mflv_2644 | proA | 0.0 | 78.55% (415) | gamma-glutamyl phosphate reductase |
| M. tuberculosis H37Rv | Rv2427c | proA | 0.0 | 83.37% (415) | gamma-glutamyl phosphate reductase |
| M. leprae Br4923 | MLBr_01458 | proA | 0.0 | 83.62% (409) | gamma-glutamyl phosphate reductase |
| M. abscessus ATCC 19977 | MAB_1618 | proA | 1e-174 | 74.48% (435) | gamma-glutamyl phosphate reductase |
| M. marinum M | MMAR_3753 | proA | 0.0 | 81.84% (435) | gamma-glutamyl phosphate reductase protein ProA |
| M. smegmatis MC2 155 | MSMEG_4584 | proA | 0.0 | 75.23% (436) | gamma-glutamyl phosphate reductase |
| M. thermoresistible (build 8) | TH_0792 | proA | 0.0 | 80.00% (415) | PROBABLE GAMMA-GLUTAMYL PHOSPHATE REDUCTASE PROTEIN PROA |
| M. ulcerans Agy99 | MUL_3696 | proA | 0.0 | 81.38% (435) | gamma-glutamyl phosphate reductase |
| M. vanbaalenii PYR-1 | Mvan_3934 | proA | 1e-180 | 77.35% (415) | gamma-glutamyl phosphate reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2644|M.gilvum_PYR-GCK MSVQAPSVP-------------------DLRQQVHDAARRARSAARGLAT
Mvan_3934|M.vanbaalenii_PYR-1 MSVQAPSVP-------------------DLRQQVHDAARRARGAARALAS
MSMEG_4584|M.smegmatis_MC2_155 MSVEAQSRSGAVD----------TQEPADLREQVHSAARRARVAARTLAT
TH_0792|M.thermoresistible__bu MSVEAPSVP-------------------GLRQQVHDAARRARGASRGLAT
MAV_1747|M.avium_104 MGLQAPFRSGGGT---SRPQGRAAPRAVDLRREVHDAARRARVASRVLAL
MLBr_01458|M.leprae_Br4923 -----------------------------MRQEVHDAARRARVAARALAV
MMAR_3753|M.marinum_M MTLQAAPRSAAA-----------QQREPDLRQEVHDAARRARVAARLLAV
MUL_3696|M.ulcerans_Agy99 MTLQAAPRSAAA-----------QQREPDLRQEVHDAARRARVAARLLAV
Mb2451c|M.bovis_AF2122/97 MTVPAPS-------------------QLDLRQEVHDAARRARVAARRLAS
Rv2427c|M.tuberculosis_H37Rv MTVPAPS-------------------QLDLRQEVHDAARRARVAARRLAS
MAB_1618|M.abscessus_ATCC_1997 MSVSTNELAPGAAGSAASAPVTEAPATADLRAGVHDAARRARVAARDLAR
:* **.****** *:* **
Mflv_2644|M.gilvum_PYR-GCK LSTDTKDRALRTAADHVLMNTRAILEANEADLDTARSSGTPEAMLDRLAL
Mvan_3934|M.vanbaalenii_PYR-1 LSTETKNRALCTAADHVLMNTRTILDANTADLDAARAAGTPEAMLDRLAL
MSMEG_4584|M.smegmatis_MC2_155 LSAEAKNRALHAAADSVLANVDAVLAANAADVDAARQGGTPEAMIDRLAL
TH_0792|M.thermoresistible__bu LGTTVKNQALHAAADAILAGADAILAANAEDLETARAAGTPEAMLDRLAL
MAV_1747|M.avium_104 LPTVAKDQALRSAADAIAGHAELILSANAEDLETARAAGTPSAMLDRLAL
MLBr_01458|M.leprae_Br4923 MPTVAKDHALRAAATAVTAHTEEILAANAEDLQAARATDTLAATLDRLAL
MMAR_3753|M.marinum_M VPTGVKDRALHAAADAILAHVDRILSANAEDLDAARAADTPAAMLDRLAL
MUL_3696|M.ulcerans_Agy99 VPTGVKDRALHAAADAILAHVDRILSANAEDLDAARAADTPAAMLDRLAL
Mb2451c|M.bovis_AF2122/97 LPTTVKDRALHAAADELLAHRDQILAANAEDLNAAREADTPAAMLDRLSL
Rv2427c|M.tuberculosis_H37Rv LPTTVKDRALHAAADELLAHRDQILAANAEDLNAAREADTPAAMLDRLSL
MAB_1618|M.abscessus_ATCC_1997 LTADAKNSALLAAADAIVASTDSIIAANADDLRRARDAGIGEAMLDRLAL
: : .*: ** :** : :: ** *: ** . * :***:*
Mflv_2644|M.gilvum_PYR-GCK TPARVEGIADGLRQVAGLPDPVGEVLRGRTLPNGLQLRQQRVPLGVVGIV
Mvan_3934|M.vanbaalenii_PYR-1 NPARVEGIADGLRQVAGLPDPIGEVIRGRTLPNGLQLRQQRVPLGVVGIV
MSMEG_4584|M.smegmatis_MC2_155 NPQRVDGIAAGLRQVAALPDPVGEVLRGKTLPNGLQLRQQRVPLGVVGMV
TH_0792|M.thermoresistible__bu NPQRIDGIAAGLRQVAGLPDPVGEVLRGRTLPNGLQLRQQRVPLGVVGIV
MAV_1747|M.avium_104 DTKRVEGIAAGLRQVAGLPDPVGEVLRGYTLPNGLQLRQQRVPLGVVGMI
MLBr_01458|M.leprae_Br4923 DRQRIDQIAGGLRQVAALPDPIGEVLRGYTLPNGLQLRQQRVPLGVVGMI
MMAR_3753|M.marinum_M NPQRVDGIAAGLRQVAGLPDPVGEVLRGYTLPNGLQLRQQRVPLGVVGMI
MUL_3696|M.ulcerans_Agy99 NPQRVDGIAAGLRQVAGLPGPVGEVLRGYTLPNGLQLRQQRVPLGVVGMI
Mb2451c|M.bovis_AF2122/97 NPQRVDGIAAGLRQVAGLRDPVGEVLRGYTLPNGLQLRQQRVPLGVVGMI
Rv2427c|M.tuberculosis_H37Rv NPQRVDGIAAGLRQVAGLRDPVGEVLRGYTLPNGLQLRQQRVPLGVVGMI
MAB_1618|M.abscessus_ATCC_1997 DTARVAGIADGLRQVAGLPDPVGEILRGSTRPNGLRLRQIRVPLGVVGMV
*: ** ******.* .*:**::** * ****:*** ********::
Mflv_2644|M.gilvum_PYR-GCK YEGRPNVTVDAFGLTLKSGNAVLLRGSSSAARSNAALVDALRAALATELL
Mvan_3934|M.vanbaalenii_PYR-1 YEGRPNVTVDAFGLTLKSGNAVLLRGSSSAARSNAALVNALRAALATEGL
MSMEG_4584|M.smegmatis_MC2_155 YEGRPNVTVDAFGLTLKSGNAALLRGSSSAARSNQALVDALRSALAEEGL
TH_0792|M.thermoresistible__bu YEGRPNVTVDAFGLTLKSGNAVLLRGSSSAARSNAALVEVLRASLTEQQL
MAV_1747|M.avium_104 YEGRPNVTVDAFGLALKSGNAVLLRGSSSATRSNQALVQVLRASLVSEDL
MLBr_01458|M.leprae_Br4923 YEGRPNITVDAFGLAFKSGNAALLRGSSSAAKSNQALVEVLHAALVSEDL
MMAR_3753|M.marinum_M YEGRPNVTVDAFGLTLKSGNAALLRGSSSAARSNEALVNVLRSALDSEQL
MUL_3696|M.ulcerans_Agy99 YEGRPNVTVDAFGLTLKSGNAALLRGSSSAARSNEALVNVLRSALDSEQL
Mb2451c|M.bovis_AF2122/97 YEGRPNVTVDAFGLTLKSGNAALLRGSSSAAKSNEALVAVLRTALVGLEL
Rv2427c|M.tuberculosis_H37Rv YEGRPNVTVDAFGLTLKSGNAALLRGSSSAAKSNEALVAVLRTALVGLEL
MAB_1618|M.abscessus_ATCC_1997 YEGRPNVTVDAFGLTLKSGNAVLLRGSSSAATSNTELVRVLRSSLVASGL
******:*******::*****.********: ** ** .*:::* *
Mflv_2644|M.gilvum_PYR-GCK DVDAVQLLPSADRASVTHLIQARGLVDVVIPRGGAGLIDAVVRDAQVPTI
Mvan_3934|M.vanbaalenii_PYR-1 DTDAVQLLPSEDRASVTHLIQARGLVDVVIPRGGAGLIDAVVRDAQVPTI
MSMEG_4584|M.smegmatis_MC2_155 PLDAVQLLPSQDRASVTHLIQARGLVDVVIPRGGAGLIDAVVRDAQVPTI
TH_0792|M.thermoresistible__bu DPDAVQLLPSDDRASVTHLIQARGLVDVVIPRGGAGLIEAVVRDAQVPTI
MAV_1747|M.avium_104 PADAVQLLSADDRATVTHLIQARGLVDVVIPRGGANLINAVVRDAQVPTI
MLBr_01458|M.leprae_Br4923 PADAVQLLSAADRSTVTHLIQARGLVDVVIPRGGANLIDAVVRDAQVPII
MMAR_3753|M.marinum_M PADAVQLLSSADRSTVTHLIQARGLVDVAIPRGGAGLIDAVVRDAQVPTI
MUL_3696|M.ulcerans_Agy99 PADAVQLLSSADRSTVTHLIQARGLVDVAIPRGGAGLIDAVVRDAQVPTI
Mb2451c|M.bovis_AF2122/97 PADAVQLLSAADRATVTHLIQARGLVDVVIPRGGAGLIEAVVRDAQVPTI
Rv2427c|M.tuberculosis_H37Rv PADAVQLLSAADRATVTHLIQARGLVDVVIPRGGAGLIEAVVRDAQVPTI
MAB_1618|M.abscessus_ATCC_1997 PADTVQLLDSRDRVTVTHLIQARGLVDVVIPRGGAGLIDAVVRDAQVPTI
*:**** : ** :*************.******.**:********* *
Mflv_2644|M.gilvum_PYR-GCK ETGVGNCHVYVHSSADLDTAESIVLNAKTRRPSVCNAAESLLIDAAVADV
Mvan_3934|M.vanbaalenii_PYR-1 ETGVGNCHVFVHESADLDMAEEIVLNAKTRRPSVCNAAESLLIDAAIADV
MSMEG_4584|M.smegmatis_MC2_155 ETGVGNCHVYVHSSADIDMAEKILLNAKTRRPSVCNAAETLLVDRALTDT
TH_0792|M.thermoresistible__bu ETGTGNCHVYVHSAADLDIAERIVLNSKTRRPSVCNSAESLLIDAGIADE
MAV_1747|M.avium_104 ETGVGNCHVYVHEGADLEVAERVLLNSKTRRPSVCNAAETLLVDAAIAEE
MLBr_01458|M.leprae_Br4923 ETGVGNCHVYVHQAADLEVAERILLNSKTRRPSVCNAAETLLIDAAIADH
MMAR_3753|M.marinum_M ETGVGNCHVYVHEAADLDVAERILLNSKTRRPSVCNAAETLLVDAAIAEH
MUL_3696|M.ulcerans_Agy99 ETGVGNCHVYVHEAADLDVAERILLNSKTRRPSVCNAAETLLVDAAIAEH
Mb2451c|M.bovis_AF2122/97 ETGVGNCHVYVHQAADLDVAERILLNSKTRRPSVCNAAETLLVDAAIAET
Rv2427c|M.tuberculosis_H37Rv ETGVGNCHVYVHQAADLDVAERILLNSKTRRPSVCNAAETLLVDAAIAET
MAB_1618|M.abscessus_ATCC_1997 ETGVGNCHVYVHADADLALAERILLNSKTRRPSVCNAAETLLIDRAIAEV
***.*****:** **: ** ::**:*********:**:**:* .:::
Mflv_2644|M.gilvum_PYR-GCK AVPRLTDALQAAGVTVH--------IDPTEDELRAEFLSMDIALAIVDGV
Mvan_3934|M.vanbaalenii_PYR-1 AVPRLTGALTAAGVTVH--------ADPSEDELRAEFLSMDIALAIVDGV
MSMEG_4584|M.smegmatis_MC2_155 ALPRLVKALQDAGVTVH--------ADPTEDELRAEFLSMDIALAVVDGL
TH_0792|M.thermoresistible__bu ALPRLVGALEQAGVVVR--------RDPSEEDLRAEFLSMDIALVVVDGL
MAV_1747|M.avium_104 ALPRLVTALQDAGVTVHGGF----ARDATEDDLRREYLSMDIAVAVVDGV
MLBr_01458|M.leprae_Br4923 TMPRLVAALQGAGVTVHGGL----SGDPDKADLCREYLSMDIAVAVVDGV
MMAR_3753|M.marinum_M AMPRLIGALQDAGVTVH--------LDADEQDLRREYLAMEIAVAVVDGV
MUL_3696|M.ulcerans_Agy99 AMPRLIGALQDAGVTVH--------LDADEQDLRREYLAMEIAVAVVDGV
Mb2451c|M.bovis_AF2122/97 ALPRLLAALQHAGVTVH--------LDPDEADLRREYLSLDIAVAVVDGV
Rv2427c|M.tuberculosis_H37Rv ALPRLLAALQHAGVTVH--------LDPDEADLRREYLSLDIAVAVVDGV
MAB_1618|M.abscessus_ATCC_1997 AVPQLVGALQAAGVTVHADLPVPGVVPVTEDDFGTEYLSLDIAVALVDGL
::*:* ** ***.*: : :: *:*:::**:.:***:
Mflv_2644|M.gilvum_PYR-GCK DGAISHINEYGTGHTEAIVTTDLAAAQRFTERVDAAAVMVNASTAFTDGE
Mvan_3934|M.vanbaalenii_PYR-1 DGAISHINEYGTGHTEAIVTTDLAAAQRFSERVDAAAVMVNASTAFTDGE
MSMEG_4584|M.smegmatis_MC2_155 DAAIDHINTYGTGHTEAIVTTDLAAAQRFTERVDAAAVMVNASTSFTDGE
TH_0792|M.thermoresistible__bu DAAIEHINEYGTGHTEAIVTTDLAAAQKFSDEVDAAAVMVNASTAFTDGE
MAV_1747|M.avium_104 DAAIAHINEYGTGHTEAIVTTNMAAAQRFTDGVDAAAVMVNASTAFTDGE
MLBr_01458|M.leprae_Br4923 DAAIAHINEYGTGHTEAIVTTNMAAAQRFADGVDSATVMVNASTAFTDGG
MMAR_3753|M.marinum_M DGAIAHINEYGTGHTEAIVTTNLAAAQRFTERIDAAAVMVNASTAFTDGE
MUL_3696|M.ulcerans_Agy99 DGAIAHINEYGTGHTEAIVTTNLAAAQRFTERIDAAAVMVNASTAFTDGE
Mb2451c|M.bovis_AF2122/97 DAAIAHINEYGTGHTEAIVTTNLDAAQRFTEQIDAAAVMVNASTAFTDGE
Rv2427c|M.tuberculosis_H37Rv DAAIAHINEYGTGHTEAIVTTNLDAAQRFTEQIDAAAVMVNASTAFTDGE
MAB_1618|M.abscessus_ATCC_1997 DAAIEHIDRYSSGHTDAIVTTNLSAAQEFTDRVDSAAVMVNASTAFTDGE
*.** **: *.:***:*****:: ***.*:: :*:*:*******:****
Mflv_2644|M.gilvum_PYR-GCK QFGFGAEIGISTQKLHARGPMGLPELTSTKWIVWGDGHTRPA
Mvan_3934|M.vanbaalenii_PYR-1 QFGFGAEIGISTQKLHARGPMGLPELTSTKWIVWGDGHTRPA
MSMEG_4584|M.smegmatis_MC2_155 QFGFGAEIGISTQKLHARGPMGLPELTSTKWIVWGDGQIRPA
TH_0792|M.thermoresistible__bu QFGFGAEIGISTQKLHARGPMGLPELTSTKWIVWGHGHTRPA
MAV_1747|M.avium_104 QFGFGAEIGISTQKLHARGPMGLPELTSTKWIAWGDGQTRPA
MLBr_01458|M.leprae_Br4923 QFGFGAEIGISTQKLHARGPMGLSEMTSTKWIVWGDGQIRPA
MMAR_3753|M.marinum_M QFGFGAEIGISTQKLHARGPMGLPELTSTKWIVWGEGHTRPA
MUL_3696|M.ulcerans_Agy99 QFGFGAEIGISTQKLHARGPMGLPELTSTKWIVRGEGHTRPA
Mb2451c|M.bovis_AF2122/97 QFGFGAEIGISTQKLHARGPMGLPELTSTKWIAWGAGHTRPA
Rv2427c|M.tuberculosis_H37Rv QFGFGAEIGISTQKLHARGPMGLPELTSTKWIAWGAGHTRPA
MAB_1618|M.abscessus_ATCC_1997 QFGFGAEIGISTQKLHARGPMGLPELTTTKWIVLGDGQIRPA
***********************.*:*:****. * *: ***