For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSDSQSVPVPDDDVDPRRIRSRTRLLDAAATLLSTGGVEAVTIDAVTKASKVARTTLYRHFHSSSHLLAA TFERLLPQVTTPAPTSGPLREQLIELLSRQAALFNDAPLHVTTLAWLSLGPTGSNNEADDRHTSGALRAR VVDQYRQPFDAILTSPKAQAELDDFDRELALCQLVGPLAFARMTGIRTITHNDCANIVDDFLTAHRKTDS DAKESKIATSRRAAPRV
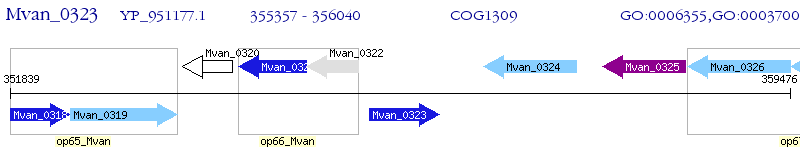
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0323 | - | - | 100% (227) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0230 | - | 2e-07 | 30.50% (141) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5312 | - | 7e-07 | 37.50% (80) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1581 | - | 1e-07 | 33.71% (89) | regulatory protein |
| M. gilvum PYR-GCK | Mflv_4579 | - | 1e-121 | 97.33% (225) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1556 | - | 1e-07 | 33.71% (89) | regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0579c | - | 5e-08 | 42.86% (70) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4834 | - | 2e-78 | 67.70% (226) | TetR family transcriptional regulator |
| M. avium 104 | MAV_3805 | - | 1e-73 | 70.20% (198) | putative transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1430 | - | 2e-43 | 46.73% (199) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2910 | - | 3e-66 | 63.27% (196) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_1934 | - | 5e-07 | 26.52% (181) | regulatory protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1581|M.bovis_AF2122/97 -------------MVGAVTQIA-----------DRPTDPSPWSPRE-TEL
Rv1556|M.tuberculosis_H37Rv -------------MVGAVTQIA-----------DRPTDPSPWSPRE-TEL
MUL_1934|M.ulcerans_Agy99 --------------MGAMTQTA-----------DRSAQVSPWSPRE-AEL
Mvan_0323|M.vanbaalenii_PYR-1 --------------MSDSQSVP---------VPDD--DVDPRRIRSRTRL
Mflv_4579|M.gilvum_PYR-GCK --------------MSDSQSVP---------VPDD--DVDPRRIRSRTRL
MAV_3805|M.avium_104 --------------MG-TQPAE---------TPDDEDDVDPRRTRSRTRL
MMAR_4834|M.marinum_M -------------MERDTLEFP---------VDDD-DDVDPRRLRSRTRL
TH_2910|M.thermoresistible__bu --------------MAAVDPAEPSARGSDDAVDDDDVDVDPRKLRSRARL
MSMEG_1430|M.smegmatis_MC2_155 --------------MTPGS-------------------NDPRPARSRARL
MAB_0579c|M.abscessus_ATCC_199 MATPRTSSASEAAQSGDGDTDATSQPAAVSALSDDDLGSSAQRERR-KRI
.. * .:
Mb1581|M.bovis_AF2122/97 LAVTLRLLQEHGYDRLTVDAVAASARASKATVYRRWPSKAELVLAAFIEG
Rv1556|M.tuberculosis_H37Rv LAVTLRLLQEHGYDRLTVDAVAASARASKATVYRRWPSKAELVLAAFIEG
MUL_1934|M.ulcerans_Agy99 LAVTLQLLQEHGYDRLTIDAVANTARASKATVYRRWPSKAELVLAAFIEG
Mvan_0323|M.vanbaalenii_PYR-1 LDAAATLLSTGGVEAVTIDAVTKASKVARTTLYRHFHSSSHLLAATFERL
Mflv_4579|M.gilvum_PYR-GCK LDAAATLLSTGGVEAVTIDAVTKASKVARTTLYRHFHSSSHLLAATFERL
MAV_3805|M.avium_104 LDAAANLLKTGGIEAVTIDAVTKASKVARTTLYRHFNSSSQLLAATFERL
MMAR_4834|M.marinum_M LDAATKLLSAGGIEAVTIDAVTKASKVARTTLYRHFSSSTQLLAATFERL
TH_2910|M.thermoresistible__bu LDAATSLLTSGGVEAVTVDAVTRLSKVARTTLYRHFESTTHLLAEAFERL
MSMEG_1430|M.smegmatis_MC2_155 LEAATVLLRSGGPSAVTVDAVTRAANVARATLYRHFPSGNDLLAAAFRQL
MAB_0579c|M.abscessus_ATCC_199 LDATLAIASKGGYEAVQMRTVAERADVAVGTLYRYFPSKVHLLVSALERE
* .: : * . : : :*: : .: *:** : * .*: :: .
Mb1581|M.bovis_AF2122/97 IRQVAVP-----PNTGNLRDDLLRLG---ELICREVGQHASTIRAVLVEV
Rv1556|M.tuberculosis_H37Rv IRQVAVP-----PNTGNLRDDLLRLG---ELICREVGQHASTIRAVLVEV
MUL_1934|M.ulcerans_Agy99 VRQVAVA-----PNTGSLRDDLVKIG---EGICGQAHQQASTIRAVLVEV
Mvan_0323|M.vanbaalenii_PYR-1 LPQVTTPA----PTSGPLREQLIELLSRQAALFNDAPLHVTTLAWLSLGP
Mflv_4579|M.gilvum_PYR-GCK LPQVTTPA----PTSGPLREQLIELLSRQAALFNDAPLHVTTLAWLSLGP
MAV_3805|M.avium_104 LPQVIPPA----PTSGTLRERLIELLSRQADLFAEAPVQVTTLAWAALGP
MMAR_4834|M.marinum_M LPQVHPP-----PATGSMRDQLIELLSRQATLFQEAPLHVTTLAWVALGP
TH_2910|M.thermoresistible__bu LPQVTSP-----PATGDLRDQLIELVHRQAALIDEAPLHLTALAWLALGP
MSMEG_1430|M.smegmatis_MC2_155 IPAAPTP-----PQEGTLRDRLIALVSAQAELIAEAPVTVTAMSWLALGA
MAB_0579c|M.abscessus_ATCC_199 FEQIDAKSDRNAVSGGTPYERLHAAITRLNRNMQRNPLLTEAMTRALVFA
. * : * :: :
Mb1581|M.bovis_AF2122/97 SRN----PALNDV----LQHQFVDHRKALIQYILQQAVDRGEISSAAISD
Rv1556|M.tuberculosis_H37Rv SRN----PALNDV----LQHQFVDHRKALIQYILQQAVDRGEISSAAISD
MUL_1934|M.ulcerans_Agy99 SRH----PALSDA----LQRQFVDQRKALIQQVLQQAVDRGEITQAAITD
Mvan_0323|M.vanbaalenii_PYR-1 TGS-NNEADDRHT-SGALRARVVDQYRQPFDAILTSPKAQAELDDFDREL
Mflv_4579|M.gilvum_PYR-GCK TGS-NNEADDRHT-SVALRARVVDQYRQPFDAILTSPKAQAELDDFDREL
MAV_3805|M.avium_104 AET-HEPNTVQHAGAGTLRMRVIEQYRRPFDEILYSPESVDQLGELDIEL
MMAR_4834|M.marinum_M TPDGTQETQDRHA----LRARIIDQYRQPFVALLQSPEARADLDDFDMEL
TH_2910|M.thermoresistible__bu DPN----SDARQP-LAGLSARVIEQYRQPFDELLAGAAARDELDDFDTTL
MSMEG_1430|M.smegmatis_MC2_155 DMEHLPLSASPEV--LSLRERVAEQYAAPFEAIFTSPEAREELGDVDRAQ
MAB_0579c|M.abscessus_ATCC_199 DAS---------------AAGEVDHVGRLMDGIFARAMAG-EDDPTDAQF
:: : :: . :
Mb1581|M.bovis_AF2122/97 ELWDLLPGYLIFRSIIPNRPPTQDTVQALVDDVILPSLTRSTG-------
Rv1556|M.tuberculosis_H37Rv ELWDLLPGYLIFRSIIPNRPPTQDTVQALVDDVILPSLTRSTG-------
MUL_1934|M.ulcerans_Agy99 ELWDLLPGYLIFRSIIPNRPPTPHTVQLLVDDFIMPGLTQATG-------
Mvan_0323|M.vanbaalenii_PYR-1 ALCQLVGPLAFARMTGIRTITHNDCAN-IVDDFLTAHRKTDSDAKESKIA
Mflv_4579|M.gilvum_PYR-GCK ALCQLVGPLAFARMTGIRTITHNDCAN-IVDDFLTAHRKTDSDAKESKIA
MAV_3805|M.avium_104 ALCQLVGPLVFARMTGLRVITHQDCTR-IVEGFIAAQTGDQPAWVEASSP
MMAR_4834|M.marinum_M ILCQLIGPVAFARLTGLRAIDRQDCER-IVDDFLVAHRRQADKPAQRAVS
TH_2910|M.thermoresistible__bu AIIQLLGPIVFAKLTGMKTLTPADRVR-IVDDFLVAHRPPTPETA-----
MSMEG_1430|M.smegmatis_MC2_155 AAALLLGPIVLGKLSTLPEFDYATCVRSAVDGFLATHRRTVSESSNESAG
MAB_0579c|M.abscessus_ATCC_199 HIARVISDVWTSNMIAWLTRRASATDVSHRLDRTVRLLLGIT--------
:: . .
Mb1581|M.bovis_AF2122/97 ------------
Rv1556|M.tuberculosis_H37Rv ------------
MUL_1934|M.ulcerans_Agy99 ------------
Mvan_0323|M.vanbaalenii_PYR-1 TSRRAAPRV---
Mflv_4579|M.gilvum_PYR-GCK TKQTGRPARLAR
MAV_3805|M.avium_104 NQ----------
MMAR_4834|M.marinum_M RKPVSRRSGSD-
TH_2910|M.thermoresistible__bu ------------
MSMEG_1430|M.smegmatis_MC2_155 A-----------
MAB_0579c|M.abscessus_ATCC_199 ------------