For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GGVTISVVRSSNPVGLAEASESMQRTIRALDDLIESRQRTLDQLSLVWSGAASDTAIRRGRRLLREKNNF RDRLHAIQTTLFRGGSQLSALREEILSTTSQATTLGGIVGDDGSVQPTSVAHMLTPTIATAYSTVLRKLL HTFEAVDEATASGLRGGRPGTGESIYQAVDFVVGGGAPEAPPDQDPAATTRRQNEINAFKQVFGRHPESP TDWKTAAALDPHSYDDKYNDEPPSVVVGRIEPVPGQGIVKSGLFIPRDVVFNVPRDDLGDNRGFDPNSGP EETRVSIYVDYENGLVIARQNPSVDTAGNVRVLTPDIKVQQAPGGAVRIQYEAANGFAPPGAELSGHVVR GDIVVTPGAGSAAASVDGVIGDYPSLEVYQDLPTGQTYTLAQDAADSGNVYGPLTELPFSHEIGEGTAAF APFESLTPQLPGGPRYPGDAMPYVPGRVDPNTPTELAPADKIPNVVVVR*
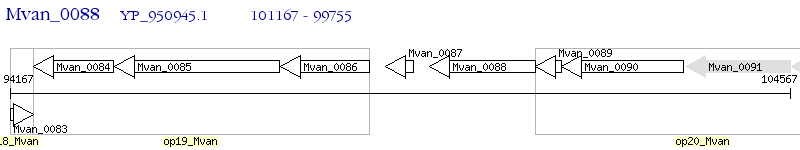
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0088 | - | - | 100% (470) | hypothetical protein Mvan_0088 |
| M. vanbaalenii PYR-1 | Mvan_1151 | - | 9e-59 | 37.44% (454) | hypothetical protein Mvan_1151 |
| M. vanbaalenii PYR-1 | Mvan_0049 | - | 6e-17 | 29.19% (209) | hypothetical protein Mvan_0049 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3088 | - | 5e-07 | 27.01% (137) | hypothetical protein Mflv_3088 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0376 | - | 3e-07 | 25.00% (152) | hypothetical protein MAB_0376 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_5295 | - | 6e-24 | 48.67% (113) | hypothetical protein MAV_5295 |
| M. smegmatis MC2 155 | MSMEG_0365 | - | 3e-15 | 24.55% (440) | hypothetical protein MSMEG_0365 |
| M. thermoresistible (build 8) | TH_2466 | - | 4e-28 | 37.63% (186) | PUTATIVE putative protein of unknown function DUF1023 |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3088|M.gilvum_PYR-GCK MGVGRLRGRRILPRRRRRSRRTVPPPVSPTVSAARRWRPDALLELAEQCD
MSMEG_0365|M.smegmatis_MC2_155 --------------------------MPGSIFEVRSSDPDALVRAGADIG
TH_2466|M.thermoresistible__bu -----------------------VWVMSATISLVEAADPQGLTAAAAQLQ
Mvan_0088|M.vanbaalenii_PYR-1 -------------------------MGGVTISVVRSSNPVGLAEASESMQ
MAB_0376|M.abscessus_ATCC_1997 --------------------------MRPTISQLRAWDVANLDAAAKAIE
MAV_5295|M.avium_104 --------------------MFAGGDPWKINATLQSGRPAQISNLAKAFH
. : .
Mflv_3088|M.gilvum_PYR-GCK DAARRLCDHIDDSVTAADASRDYWTGAAADAARGAARGIAADAARAAHRL
MSMEG_0365|M.smegmatis_MC2_155 NRAGRLQAQMDRQQAVIDDLRRGWNGASADAAIESAERSLQHMRRLHDSL
TH_2466|M.thermoresistible__bu TKXTHLDTIITAERSALAALAGGWEGTAADAALARGEQHLAQQEQLRTRL
Mvan_0088|M.vanbaalenii_PYR-1 RTIRALDDLIESRQRTLDQLSLVWSGAASDTAIRRGRRLLREKNNFRDRL
MAB_0376|M.abscessus_ATCC_1997 DASKNLDLSIDGVVRDLDSAESFWKGKAFQSAYDQASNEQSSTNRLSFAI
MAV_5295|M.avium_104 DAGRSTGEAEAAFNLARGRFEQAWTHENDENPINGSAEVQRTTTSLGVQA
* : . .
Mflv_3088|M.gilvum_PYR-GCK VNASAAARDGATQIGAARDDAVGQVDDALEHGFLVADDGTVTVGADPDPL
MSMEG_0365|M.smegmatis_MC2_155 TTLQSALQTGGADLAQQRDRIVNHVDQLHAQGWQVADDGTVSIRPGS---
TH_2466|M.thermoresistible__bu TSLQATLADAGANLSALRTRILDVAAQARSLGGLVRDDGTVIALTSG---
Mvan_0088|M.vanbaalenii_PYR-1 HAIQTTLFRGGSQLSALREEILSTTSQATTLGGIVGDDGSVQPTSVA---
MAB_0376|M.abscessus_ATCC_1997 SNVAEAVRGGHSALTAARDKVMLLVDEATSKGLHVADDGTVTAPEGN---
MAV_5295|M.avium_104 AQLPKIGADLEKVAAALAEAQRSGRGEISSLEGALQDLDDQVGEAVELEK
: : * .
Mflv_3088|M.gilvum_PYR-GCK LVALAGGRDTLAADLLQLRADELSVRIPAALDRLGAADSDAAADLVRALT
MSMEG_0365|M.smegmatis_MC2_155 VLDFFAKVSPVQRVMLEALAARASAELKAMLAQFDATDVTVGAQIRDAVA
TH_2466|M.thermoresistible__bu ------------GLMTPDVAAAYTTTLKTLLQTFDRVDQQSSQSVIQALG
Mvan_0088|M.vanbaalenii_PYR-1 ------------HMLTPTIATAYSTVLRKLLHTFEAVDEATASGLRGGRP
MAB_0376|M.abscessus_ATCC_1997 -------------RELQAEARRLTDAIRQALKSLEEADRTSSNDLRKASA
MAV_5295|M.avium_104 NPQLTASERQLLDHYIDGLEKHAIDDTKASVANLTRIRDQYSQQLHTSMA
: : . : .
Mflv_3088|M.gilvum_PYR-GCK DTDRPVTPAPTVPAGAWPVPSGDVVDGWPAMSQDRIAAQVAAMTDDQRAR
MSMEG_0365|M.smegmatis_MC2_155 G----LTAHGPAPASPPPNNAKALVDRLAGMTPDERREFLAGLSPETVHE
TH_2466|M.thermoresistible__bu DG-------ADTGAEAPPEPDIDDLKPPEGATDTEVNQWWDGLSEEDRQR
Mvan_0088|M.vanbaalenii_PYR-1 GTG---------------ESIYQAVDFVVGGGAPEAPPDQDPAATTRRQN
MAB_0376|M.abscessus_ATCC_1997 S-----------------------VDAEHNSASTQPAAFMPASSGGAASG
MAV_5295|M.avium_104 NLR-----------------EQDGYDPPIQAFDGDVPE--SPSQEQRRHN
. .
Mflv_3088|M.gilvum_PYR-GCK LIAEFPHQVGNTDGVPWDMRVAANRTNIATAILAESAADAPSRRRIAFYR
MSMEG_0365|M.smegmatis_MC2_155 MVLADPETMGNTDGVPFETRIEANDINIRNALEEELAKPSPDQARVQQLQ
TH_2466|M.thermoresistible__bu LIHERPEWIGNLDGVPASARDQANRNRLEG-LRDDPGLTVEERDTIAAID
Mvan_0088|M.vanbaalenii_PYR-1 EINAFKQVFGRHPESPTDWKTAAALDPHSYDDKYNDEPPSVVVGRIEPVP
MAB_0376|M.abscessus_ATCC_1997 SMKAGS---GGFSGGGFGGGSGGGHG------------------------
MAV_5295|M.avium_104 QIEAFKQVFGREPASAADWETAAALDSQTYDSKFNGAKSEIRVVRIRPVP
: * .
Mflv_3088|M.gilvum_PYR-GCK SLLGEVD------DPSGD-TRIARQILAFDP-GRASLVEL-HGDLG-TAR
MSMEG_0365|M.smegmatis_MC2_155 AMLTPIK------DPAGSGKLVPRKFVAFSPDGNGRMIEM-IGEIKPGIK
TH_2466|M.thermoresistible__bu DALEGEERQLLVLDFDGEHPKVAVALGDVETADHVSVYVPGTGSVTYDKP
Mvan_0088|M.vanbaalenii_PYR-1 GQG-----------IVKSGLFIPRDVVFNVPRDDLGDNRGFDPNSGPEET
MAB_0376|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_5295|M.avium_104 GQG------------------------------------VVRVSQWIEQR
Mflv_3088|M.gilvum_PYR-GCK SVAVLVPGVNTTVEGSAANAATARRFVSGSDGRLAMITYLGGPFPQVHDP
MSMEG_0365|M.smegmatis_MC2_155 GVGVVVPGTNTNLNGSGSNHTSAVELSKQSGS--PVFLYMDGDFPQGLD-
TH_2466|M.thermoresistible__bu DAGNDLPTYVEQSGWLKAETERVLEQAGRSDETVATVAWLGYEPPANVA-
Mvan_0088|M.vanbaalenii_PYR-1 RVSIYVDYENGLVIARQNPSVDTAGNVRVLTPDIKVQQAPGGAVRIQYE-
MAB_0376|M.abscessus_ATCC_1997 -------GGFGGGHGGGGGGGHSVPDGSYSNTGAPSLRPSGPSVFMNLD-
MAV_5295|M.avium_104 DVRSFPPWKRDLGNNRGANPNFDPEDTKVRCADSPTRHRLWTSHRHPFCA
Mflv_3088|M.gilvum_PYR-GCK VTAVAAADRRYALEMAPRLVAFSEDVARRV---PDSVLVTVVGHSYGGSI
MSMEG_0365|M.smegmatis_MC2_155 ----KAADPAYAASMAPKLVSFGHEIDREVGIGAPDTPVTYIGHSYGGAI
TH_2466|M.thermoresistible__bu ----QAGSPHYADDNAPKLASFINGLNSSR---DTDPNLTVLGHSYGSLV
Mvan_0088|M.vanbaalenii_PYR-1 ----------AANGFAPPGAELSGHVVRGD---------IVVTPGAGSAA
MAB_0376|M.abscessus_ATCC_1997 -----------AGQMEVARKIIEEGLRRGLS-----PEAIQIALATALTE
MAV_5295|M.avium_104 P--------SAHRQCSSRLITKSGRLVMRSS----AREPFFDAEAWRIAT
: .
Mflv_3088|M.gilvum_PYR-GCK VGTAETLGLTSDRTVFLAAAGAGVGVDDPGDWHNRNPHVQRYSMTAPGDF
MSMEG_0365|M.smegmatis_MC2_155 VGTAEQMGLRADRVLHASSAGTGILS---GDYTNPNPNVQRFSMTAPGDP
TH_2466|M.thermoresistible__bu ASEALQRGTAADNVVFMGSPGLETNV---WEWHK-NVTPEKLHLSEGHVF
Mvan_0088|M.vanbaalenii_PYR-1 ASVDGVIGDYPSLEVYQDLPTG-------QTYTLAQDAADSGNVYGPLTE
MAB_0376|M.abscessus_ATCC_1997 SGLRSLANSSVPDSMMFPNDGVG------HDHDSVGPFQQRQSWGATADL
MAV_5295|M.avium_104 VSAPFVVQGVLALLMLTLWLLGE------GPFKTHSAYAGERAWMVTASL
. :
Mflv_3088|M.gilvum_PYR-GCK IELVQGIP------------------------------GGPHGADPDEMD
MSMEG_0365|M.smegmatis_MC2_155 ISLVQSLPRDVAISSIPGVDSIPGIPHNTDGRIGNPLGGLPSATDPDNIP
TH_2466|M.thermoresistible__bu VQHADNDIVAN--------------------------LGRYGAVIPSELP
Mvan_0088|M.vanbaalenii_PYR-1 LPFSHEIG-----------------------------EGTAAFAPFESLT
MAB_0376|M.abscessus_ATCC_1997 MDPTKSAG--------------------------------KFYDQLVKVS
MAV_5295|M.avium_104 IASAVSLS----------------------------------------IS
: :
Mflv_3088|M.gilvum_PYR-GCK GVIGLPTGFYDDG-----GLVAGWEAHSGMLNR-PSGAWRTILDVLTGDP
MSMEG_0365|M.smegmatis_MC2_155 GVTRLDTGYYGAEGAHPNQVIVGPDGHGKYWDDYDSDAFKNMVAVISGGE
TH_2466|M.thermoresistible__bu GFIDLSTNAESTP----LGPRDASSGHSEYSFINKTALYNQAVVVAGLGQ
Mvan_0088|M.vanbaalenii_PYR-1 PQLPGGPRYPGDA----MPYVPGRVDPNTPTELAPADKIPNVVVVR----
MAB_0376|M.abscessus_ATCC_1997 GWQDMSVAQAAQS--------VQRSAFPDAYAKYEAQAAQIFHQVTGR--
MAV_5295|M.avium_104 VPLAWTPSSRRRG--------ISLSLAGCSALVLVGGVLYAYLTMR----
:
Mflv_3088|M.gilvum_PYR-GCK GTLSSASSIPAGG-------------------------------------
MSMEG_0365|M.smegmatis_MC2_155 ATGYVERGIETNYVDIDLGDDGDFRDEAADQAKAGAAPKAPRLPWDDNGT
TH_2466|M.thermoresistible__bu DDRFIRVED-----------------------------------------
Mvan_0088|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0376|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_5295|M.avium_104 --------------------------------------------------
Mflv_3088|M.gilvum_PYR-GCK --------------------------
MSMEG_0365|M.smegmatis_MC2_155 YERRQHPWDNPRVTDNPGLGPRVPVR
TH_2466|M.thermoresistible__bu --------------------------
Mvan_0088|M.vanbaalenii_PYR-1 --------------------------
MAB_0376|M.abscessus_ATCC_1997 --------------------------
MAV_5295|M.avium_104 --------------------------