For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEVHAEPAAPTLRDLATAGLGLSVPDGTAQELDRPVSWVHTTEMRDPSRYLRGGELVCTVGISLHTDQDC ATFVDALSRARAAGVCFGVGDEHDAVPEALLAQCRLHGLPLLVAPPSVPFSVVSRFVAEYWLGSEIAVAR ATNALVPELLSSLRRHDSPRQLLDSAGQLLGCYFLLEPEGGADSDDDAVTVPVPGLGTLVWVGRGDHPEP ALLDLIARFVRAAQGERDIEAALARERVGQLLSLVERRMLLPDALSQLLEWPGFSAGEIACSAWPAGAGA LVSMAFPDALIGDAPDLCLMLTVGALEVTDDLALPTGHSAPVPLTDIGSAIAQARIALTLAEHRGGRVGP DQLSTLESLLEQLPLAQLAPFKQLLIAPLAELDSQRGTQHVRTLRVFLACNGSLSETAKELFLHTNTVRH RLSRIQEITGRDPLQHNDLTAFVIGLWAADRAERPAGL*
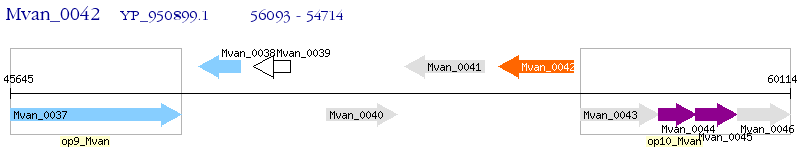
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0042 | - | - | 100% (459) | purine catabolism PurC domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1854 | - | 1e-20 | 26.54% (486) | purine catabolism PurC domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1825 | - | 5e-09 | 31.08% (148) | purine catabolism PurC domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2266 | - | 1e-09 | 36.08% (97) | hypothetical protein Mb2266 |
| M. gilvum PYR-GCK | Mflv_0801 | - | 0.0 | 83.74% (455) | transcriptional regulator, CdaR |
| M. tuberculosis H37Rv | Rv2242 | - | 2e-09 | 36.08% (97) | hypothetical protein Rv2242 |
| M. leprae Br4923 | MLBr_01652 | - | 2e-08 | 40.62% (64) | hypothetical protein MLBr_01652 |
| M. abscessus ATCC 19977 | MAB_0825 | - | 8e-17 | 26.55% (516) | peptidase |
| M. marinum M | MMAR_3335 | - | 2e-09 | 31.62% (136) | hypothetical protein MMAR_3335 |
| M. avium 104 | MAV_2195 | - | 1e-09 | 32.85% (137) | hypothetical protein MAV_2195 |
| M. smegmatis MC2 155 | MSMEG_1068 | - | 1e-172 | 67.03% (461) | regulatory protein |
| M. thermoresistible (build 8) | TH_1338 | - | 2e-08 | 42.25% (71) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1307 | - | 3e-09 | 42.19% (64) | hypothetical protein MUL_1307 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3335|M.marinum_M MNDNQFS-AIARLGSARSPLELLETVPDSLLRRLKQYSGRLATEAVSAMQ
MUL_1307|M.ulcerans_Agy99 MNNNQFS-AIARLGSARSPLELLETVPDSLLRRLKQYSGRLATEAVSAMQ
Mb2266|M.bovis_AF2122/97 MNDNQLA-PVARP---RSPLELLDTVPDSLLRRLKQYSGRLATEAVSAMQ
Rv2242|M.tuberculosis_H37Rv MNDNQLA-PVARP---RSPLELLDTVPDSLLRRLKQYSGRLATEAVSAMQ
MLBr_01652|M.leprae_Br4923 MNHNHFV-PAPPR---RLPLELLDTVPDSLLRRLKQYSGRLATEAVTAMQ
MAV_2195|M.avium_104 MNDNPFAGPFAKHP--RSPLETLDTVPESVLRRLKQYSGRLATEAVSVMQ
TH_1338|M.thermoresistible__bu VPDNRFV----PP---QSTLEVLESVPESVLRRLKQYSGRLATEAVHTME
Mvan_0042|M.vanbaalenii_PYR-1 MSEVHAEPAAPTLRDLATAG----LGLSVPDGTAQELDRPVSWVHTTEMR
Mflv_0801|M.gilvum_PYR-GCK MSEDRAESAAPTLRDLLEAAQ---LSLSGPDPADRNLDREVSWVHTTEMR
MSMEG_1068|M.smegmatis_MC2_155 MPD--RDDVAPTLRDLLNSHLGVVAAQNLTDDLSAELDRTITWAHTTELR
MAB_0825|M.abscessus_ATCC_1997 MAEYRLTVPITVRDLTQRADLGLVLSAG-----EAGAARAIKWAHAIELN
: . . : . :.
MMAR_3335|M.marinum_M ERLPFFADLEASQRASVAL-VVQTAVVNFAEWMQNPRSD--VSYTAQAFE
MUL_1307|M.ulcerans_Agy99 ERLPFFADLEASQRASVAL-VVQTAVVNFAEWMQNPRSD--VSYTAQAFE
Mb2266|M.bovis_AF2122/97 ERLPFFADLEASQRASVAL-VVQTAVVNFVEWMHDPHSD--VGYTAQAFE
Rv2242|M.tuberculosis_H37Rv ERLPFFADLEASQRASVAL-VVQTAVVNFVEWMHDPHSD--VGYTAQAFE
MLBr_01652|M.leprae_Br4923 ERLPFFADLEASQRASVTL-VVQTAVVNFVQWMQNPHSD--VSYTAQAFE
MAV_2195|M.avium_104 DRLPFFADLEASQRASVSL-VVQTAVVNFAEWMQDPKSN--VRHTAQAFE
TH_1338|M.thermoresistible__bu ERLPFFADLEASQRASVHL-VVQTAVVNFVEWMRDPRSN--VTYTMQAFE
Mvan_0042|M.vanbaalenii_PYR-1 DPSRYLRGGELVCTVGISL-HTDQDCATFVDALSRARAAGVCFGVGDEHD
Mflv_0801|M.gilvum_PYR-GCK DPSRYLRGGELVCTVGISL-HSAEDCSTFVDALAGAGAAGVCFGVGDGHD
MSMEG_1068|M.smegmatis_MC2_155 DPSRYLRGGELVCTVGISL-QTPDDCEAFVAALCESGAAGLCFGTGDAHD
MAB_0825|M.abscessus_ATCC_1997 DPTPYLFGGELVMTTGLTVGLSHAEQYEYAARLSGKNVAALAFDTGTTFS
: :: . * ..: : :. : . ..
MMAR_3335|M.marinum_M LVPQDLTRRIALR--------------------------HTVDMVRVTME
MUL_1307|M.ulcerans_Agy99 LVPQDLTRRIALR--------------------------HTVDMVRVTME
Mb2266|M.bovis_AF2122/97 LVPQDLTRRIALR--------------------------QTVDMVRVTME
Rv2242|M.tuberculosis_H37Rv LVPQDLTRRIALR--------------------------QTVDMVRVTME
MLBr_01652|M.leprae_Br4923 LVPQDLARRIALR--------------------------HTVDMVRVTME
MAV_2195|M.avium_104 LVPQDLARRIALR--------------------------HSVEMVRVTME
TH_1338|M.thermoresistible__bu VVPQDLTRRIALR--------------------------QTVEMVRTTME
Mvan_0042|M.vanbaalenii_PYR-1 AVPEALLAQCRLHGLPLLVAPPSVPFSVVSRFVAEYWLGSEIAVARATNA
Mflv_0801|M.gilvum_PYR-GCK DVPAALLDACRRAGLPVLVAPPSVPFSVISRFVAESRLGTEIAVARATNA
MSMEG_1068|M.smegmatis_MC2_155 AVPDALVAACTRRRLPLLIAPPNVLFATVSRYVADFQVGGELAVARATNT
MAB_0825|M.abscessus_ATCC_1997 EVPPGILAAGDELGLPILQVPVSTPFIAITRAVIDELTADELRAVQRVVD
** : : .: .
MMAR_3335|M.marinum_M FFEEVVPLLARSEEQ--------------------LTALTVG--------
MUL_1307|M.ulcerans_Agy99 FFEEVVPLLARSEEQ--------------------LTALTVG--------
Mb2266|M.bovis_AF2122/97 FFEEVVPLLARSEEQ--------------------LTALTVG--------
Rv2242|M.tuberculosis_H37Rv FFEEVVPLLARSEEQ--------------------LTALTVG--------
MLBr_01652|M.leprae_Br4923 FFEEVVPLLARSEEQ--------------------LTALTVG--------
MAV_2195|M.avium_104 FFEEVVPLLARSEEQ--------------------LTALTVG--------
TH_1338|M.thermoresistible__bu FFEQVVPLLARSEEQ--------------------VTALTAG--------
Mvan_0042|M.vanbaalenii_PYR-1 LVPELLSSLRRHDSP--------------------RQLLDSAGQLLGCYF
Mflv_0801|M.gilvum_PYR-GCK LVPELLLSLRRHDSP--------------------RRLLDAAGQVLGCYF
MSMEG_1068|M.smegmatis_MC2_155 LVPELLASARRRESP--------------------RQLLDRAGEVLGCYF
MAB_0825|M.abscessus_ATCC_1997 QQDVLVRATLRDGAPGVVSALSRSLAATVIVTATDGTLLAEAGHDAEHLA
:: * * .
MMAR_3335|M.marinum_M ILKYSRDLAFTAASAYADAAEARG--TWDSR----------------MEA
MUL_1307|M.ulcerans_Agy99 ILKYSRDLAFTAASAYADAAEARG--TWDSR----------------MEA
Mb2266|M.bovis_AF2122/97 ILKYSRDLAFTAATAYADAAEARG--TWDSR----------------MEA
Rv2242|M.tuberculosis_H37Rv ILKYSRDLAFTAATAYADAAEARG--TWDSR----------------MEA
MLBr_01652|M.leprae_Br4923 ILKYSRDLAFTAATAYADAAEARG--TWDSR----------------MEA
MAV_2195|M.avium_104 VLKYSRDLAFTAATAYADAAEARG--TWDSR----------------MEA
TH_1338|M.thermoresistible__bu ILRYSRDLAFAAATAYADAAEARG--AWDTR----------------MEA
Mvan_0042|M.vanbaalenii_PYR-1 LLEPEGGAD-SDDDAVTVPVPGLGTLVWVGRGD-------------HPEP
Mflv_0801|M.gilvum_PYR-GCK LLEPEGAAK-GDEAAVTVPVPGLGALVWMGRGD-------------RPEP
MSMEG_1068|M.smegmatis_MC2_155 LLEPGAPADTGGPAAHTVTIDGLGTLVWVGSGT-------------PPDD
MAB_0825|M.abscessus_ATCC_1997 EITRGAFPNHSRKHASRVVADRDGYCTIQTVRAGKAERGYLAVRSEDALT
: * * .
MMAR_3335|M.marinum_M SVVDAVVRGDTGPELLS-RAAALNWDTTAPATVVVGTPASARDGS--DG-
MUL_1307|M.ulcerans_Agy99 SVVDAVVRGDTGPELLS-RAAALNWDTTAPATVVVGTPASARDGS--DG-
Mb2266|M.bovis_AF2122/97 SVVDAVVRGDTGPELLS-RAAALNWDTTAPATVLVGTPAPGPNGSNSDGD
Rv2242|M.tuberculosis_H37Rv SVVDAVVRGDTGPELLS-RAAALNWDTTAPATVLVGTPAPGPNGSNSDGD
MLBr_01652|M.leprae_Br4923 SVVDAVVRGDTGPELLS-RAAALNWDTTAPATVVVGTPTPNHDGPNGQVS
MAV_2195|M.avium_104 SVVDAVVRGDTGPELLS-RAAALNWDTTAPATVVVGTPAPGRDGSTGPGD
TH_1338|M.thermoresistible__bu NVVDAVVRGAVGPELQS-QAAALNWDATAPATVVVGLPRPD--------R
Mvan_0042|M.vanbaalenii_PYR-1 ALLDLIARFVRAAQGERDIEAALARERVGQLLSLVERRMLLPDALS--QL
Mflv_0801|M.gilvum_PYR-GCK ALLDLIARFVRAAQGERDIEAALTRERVGQLLSLVERRMLLPDALS--QL
MSMEG_1068|M.smegmatis_MC2_155 AVLDVIVRFVHAAQGERDIEAALARERVGQLLSLVERRMLLPDALN--QL
MAB_0825|M.abscessus_ATCC_1997 AAQRLLVSHAMALIAIELEKPARVADAEQRLRGAVTRALLADPAAADRSV
:. . .* : *
MMAR_3335|M.marinum_M SERAIQDVRDAAAQHGRAALTDVHG-------------TWLVAIVSGHLS
MUL_1307|M.ulcerans_Agy99 SERAIQDVRDAAAQHGRAALTDVHG-------------TWLVAIVSGHLS
Mb2266|M.bovis_AF2122/97 SERASQDVRDTAARHGRAALTDVHG-------------TWLVAIVSGQLS
Rv2242|M.tuberculosis_H37Rv SERASQDVRDTAARHGRAALTDVHG-------------TWLVAIVSGQLS
MLBr_01652|M.leprae_Br4923 SERASQEVREIAARHGRAALTDVHG-------------TWLVAIISGQLA
MAV_2195|M.avium_104 SERASQHVRDIAAQHGRAALTDVHG-------------TWLVAIVSGQLS
TH_1338|M.thermoresistible__bu LEVVSQDVHHVANRHGRTALSDVHG-------------TWLVTIVSGALS
Mvan_0042|M.vanbaalenii_PYR-1 LEWPGFSAGEIACSAWPAGAGALVS-------------MAFPDALIGDAP
Mflv_0801|M.gilvum_PYR-GCK VQWPGF-AGEIACSAWPAGAGALLS-------------ISFPGALIGDAP
MSMEG_1068|M.smegmatis_MC2_155 LDWPGLAAREVACSAWPAGAGALLS-------------MAFPTALVGDAP
MAB_0825|M.abscessus_ATCC_1997 LRYFGFDPDTEVVVVLLCNVGSVLPGELHIDRQLARGGPYLMASVSGEIV
. : : : *
MMAR_3335|M.marinum_M PTDKVFKDLLEAFSDGP--------VVIGPTAPMLTAAYHSASEAISGMN
MUL_1307|M.ulcerans_Agy99 PTDKVFKDLLEAFSDGP--------VVIGPTAPMLTAAYHSASEAISGMN
Mb2266|M.bovis_AF2122/97 PTEKFLKDLLAAFADAP--------VVIGPTAPMLTAAHRSASEAISGMN
Rv2242|M.tuberculosis_H37Rv PTEKFLKDLLAAFADAP--------VVIGPTAPMLTAAHRSASEAISGMN
MLBr_01652|M.leprae_Br4923 PTDKFFSDLLHAFSDGP--------VVIGPTAPMLTAAYHSASEAVSGMN
MAV_2195|M.avium_104 PTDKFLGDLLEAFSDGP--------VVVGPTAPMLTAAYHSASEAISGMN
TH_1338|M.thermoresistible__bu PTDRFLTDLLDVFADGP--------VVLGPTAPTLSAAYRSATEAIAGMN
Mvan_0042|M.vanbaalenii_PYR-1 DLCLMLTVGALEVTDDL-------ALPTGHSAPVPLTDIGSAIAQARIAL
Mflv_0801|M.gilvum_PYR-GCK DLCMMLTAGAVEMPDDL-------ALPTGHSAPVTLADLGSAIAQARIAL
MSMEG_1068|M.smegmatis_MC2_155 EVCLVLTDPATSATADLPAIAAELSLPSGHSATAPLAELGSAITQARIAL
MAB_0825|M.abscessus_ATCC_1997 VVLAAKNVGRASELHRSLGTALGRTICAGISRRVRLDEVPLGLHQAR-SV
: * : .
MMAR_3335|M.marinum_M AVAGWRG---APRPVLARELLPERALMGDASAIVALHTDVMRPLAD-AGP
MUL_1307|M.ulcerans_Agy99 AVAGWRG---ASRPVLARELLPERALMGDASAIVALHTDVMRPLAD-AGP
Mb2266|M.bovis_AF2122/97 AVAGWRG---APRPVLARELLPERALMGDASAIVALHTDVMRPLAD-AGP
Rv2242|M.tuberculosis_H37Rv AVAGWRG---APRPVLARELLPERALMGDASAIVALHTDVMRPLAD-AGP
MLBr_01652|M.leprae_Br4923 AVAGWSG---APRPVQARELLPERALMGDASAIVALHTDVMLPLAD-AGP
MAV_2195|M.avium_104 AVGGWRG---APRPVLARELLPERALMGDASAIVALHTDVMGPLAD-AGP
TH_1338|M.thermoresistible__bu AVAGWSG---APRPVPARELLPERALLGDQTAIAALESEVMRPLAD-AGP
Mvan_0042|M.vanbaalenii_PYR-1 TLAEHRGGRVGPDQLSTLESLLEQLPLAQLAPFKQLLIAPLAELDSQRGT
Mflv_0801|M.gilvum_PYR-GCK TLAEHRGGRVGPDQLSTLESLLEQLPLAQLAPFRQLLIDPLTELDTSRGT
MSMEG_1068|M.smegmatis_MC2_155 DLARQHGGSIGPDQLSTFDSLLEGLPLSRLTPFKQQLIDPLAESDHERGT
MAB_0825|M.abscessus_ATCC_1997 ARAHPDGRIAEFDELDIYGVLLGGRDRGELAVLAQSVAPLLGTGAD---T
. * : * . : : : .
MMAR_3335|M.marinum_M TLIETLDAYLDCGGAIEACARKLFVHPNTIRYRLKRITDFTGRDPTQPRD
MUL_1307|M.ulcerans_Agy99 TLIETLDAYLDCGGAIEACARKLFVHPNTIRYRLKRITDFTGRDPTQPRD
Mb2266|M.bovis_AF2122/97 TLIETLDAYLDCGGAIEACARKLFVHPNTVRYRLKRITDFTGRDPTQPRD
Rv2242|M.tuberculosis_H37Rv TLIETLDAYLDCGGAIEACARKLFVHPNTVRYRLKRITDFTGRDPTQPRD
MLBr_01652|M.leprae_Br4923 TLIETLDAYLDCGGAIEACARKLFVHPNTVRYRLKRITDFTGHDPTLPRD
MAV_2195|M.avium_104 TLIETLDAFLDSGGAIEACARKLFVHPNTVRYRLKRITDFTGRDPMQPRD
TH_1338|M.thermoresistible__bu TLTETLDAFLESGGAIEACARRLFVHPNTVRYRLRRIAEITGRDPAVPRD
Mvan_0042|M.vanbaalenii_PYR-1 QHVRTLRVFLACNGSLSETAKELFLHTNTVRHRLSRIQEITGRDPLQHND
Mflv_0801|M.gilvum_PYR-GCK QHVRTLRTFVACNGSLSETAKELFLHTNTVRHRLGRIQEITGRDPLQHND
MSMEG_1068|M.smegmatis_MC2_155 QHVRTLRVFLATNGSLIDTARELFLHTNTVRHRLARIHEITGRNPLHFED
MAB_0825|M.abscessus_ATCC_1997 TLLDTLESFLACNGQMETAALNLGVHRHTMRNRIARIAALLDCDLHSADT
** :: .* : * .* :* :*:* *: ** : . :
MMAR_3335|M.marinum_M AYVLRVAATVGQLNYPTPPSGAANNTITPVPLPVRRGTEGP---
MUL_1307|M.ulcerans_Agy99 AYVLRVAATVGQLNYPTPPSGAANNTITPVPLPVRRGTEGP---
Mb2266|M.bovis_AF2122/97 AYVLRVAATVGQLNYPTPH-------------------------
Rv2242|M.tuberculosis_H37Rv AYVLRVAATVGQLNYPTPH-------------------------
MLBr_01652|M.leprae_Br4923 AYVLRVAATVGKLNYPTHH-------------------------
MAV_2195|M.avium_104 AYVLRVAATVGQLNYPTHPPSAAGAAMPAVPLPVNGAARGQSGG
TH_1338|M.thermoresistible__bu AYVLRVAVSVSRLTRQSYQSSTVNPDLGSTAGWRGQAAGGHRAM
Mvan_0042|M.vanbaalenii_PYR-1 LTAFVIGLWAADRAERPAGL------------------------
Mflv_0801|M.gilvum_PYR-GCK LTTFVIGMWAADRGE-----------------------------
MSMEG_1068|M.smegmatis_MC2_155 QAAFAIGLRASDRHSRMTRRPN----------------------
MAB_0825|M.abscessus_ATCC_1997 RAALWIAIRARALLG-----------------------------
.: :. .