For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PDRDDVAPTLRDLLNSHLGVVAAQNLTDDLSAELDRTITWAHTTELRDPSRYLRGGELVCTVGISLQTPD DCEAFVAALCESGAAGLCFGTGDAHDAVPDALVAACTRRRLPLLIAPPNVLFATVSRYVADFQVGGELAV ARATNTLVPELLASARRRESPRQLLDRAGEVLGCYFLLEPGAPADTGGPAAHTVTIDGLGTLVWVGSGTP PDDAVLDVIVRFVHAAQGERDIEAALARERVGQLLSLVERRMLLPDALNQLLDWPGLAAREVACSAWPAG AGALLSMAFPTALVGDAPEVCLVLTDPATSATADLPAIAAELSLPSGHSATAPLAELGSAITQARIALDL ARQHGGSIGPDQLSTFDSLLEGLPLSRLTPFKQQLIDPLAESDHERGTQHVRTLRVFLATNGSLIDTARE LFLHTNTVRHRLARIHEITGRNPLHFEDQAAFAIGLRASDRHSRMTRRPN*
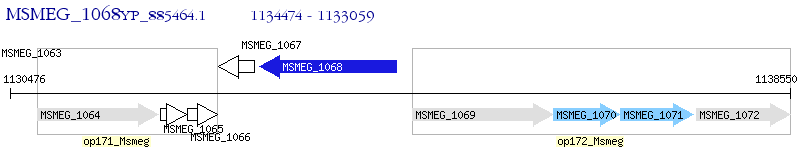
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1068 | - | - | 100% (471) | regulatory protein |
| M. smegmatis MC2 155 | MSMEG_2971 | - | 8e-26 | 26.67% (495) | regulatory protein |
| M. smegmatis MC2 155 | MSMEG_4370 | - | 2e-18 | 36.20% (163) | regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1218c | - | 5e-07 | 26.47% (204) | hypothetical protein Mb1218c |
| M. gilvum PYR-GCK | Mflv_0801 | - | 1e-162 | 64.35% (460) | transcriptional regulator, CdaR |
| M. tuberculosis H37Rv | Rv1186c | - | 6e-07 | 26.47% (204) | hypothetical protein Rv1186c |
| M. leprae Br4923 | MLBr_01652 | - | 3e-06 | 42.86% (56) | hypothetical protein MLBr_01652 |
| M. abscessus ATCC 19977 | MAB_4202c | - | 3e-19 | 26.41% (462) | putative regulatory protein |
| M. marinum M | MMAR_4254 | - | 2e-08 | 27.27% (297) | hypothetical protein MMAR_4254 |
| M. avium 104 | MAV_1330 | - | 2e-11 | 27.08% (288) | hypothetical protein MAV_1330 |
| M. thermoresistible (build 8) | TH_1338 | - | 6e-09 | 39.56% (91) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0931 | - | 5e-08 | 26.94% (297) | hypothetical protein MUL_0931 |
| M. vanbaalenii PYR-1 | Mvan_0042 | - | 1e-172 | 67.03% (461) | purine catabolism PurC domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0801|M.gilvum_PYR-GCK --MSEDRAESAAPTLRDLLEAAQ---LSLSGPDPADRNLDREVSWVHTTE
Mvan_0042|M.vanbaalenii_PYR-1 --MSEVHAEPAAPTLRDLATAG----LGLSVPDGTAQELDRPVSWVHTTE
MSMEG_1068|M.smegmatis_MC2_155 --MPD--RDDVAPTLRDLLNSHLGVVAAQNLTDDLSAELDRTITWAHTTE
Mb1218c|M.bovis_AF2122/97 MRIAGVGLGQLLLALDATVVSLVDAPRGLDLPVASTALIDSDDVRLGLAA
Rv1186c|M.tuberculosis_H37Rv MRIAGVGLGQLLLALDATVVSLVDAPRGLDLPVASTALIDSDDVRLGLAA
MMAR_4254|M.marinum_M MGVAGVGLGQLLLALDATLVSLVEAPRGLDLPVASAALVDSDDVRLGLAA
MUL_0931|M.ulcerans_Agy99 MGVAGVGLGQLLLALDATLVSLVEAPRGLDLPVASAALVDSDDVRLGLAA
MAV_1330|M.avium_104 --MAGVGLGQLLLALDATMVSLVDAPRGLDQPVASAALIDSDDVRLGLAA
MLBr_01652|M.leprae_Br4923 -------------MNHNHFVPAPPRRLPLELLDTVPDSLLR---------
TH_1338|M.thermoresistible__bu -------------VPDNRFVPPQS---TLEVLESVPESVLR---------
MAB_4202c|M.abscessus_ATCC_199 ----------MRWVLDQPELKLTLKSGAAGVGREINLALTTELA------
:
Mflv_0801|M.gilvum_PYR-GCK MR-------------DPSRYLRGGELVCTVGISLHSA-EDCSTFVDALAG
Mvan_0042|M.vanbaalenii_PYR-1 MR-------------DPSRYLRGGELVCTVGISLHTD-QDCATFVDALSR
MSMEG_1068|M.smegmatis_MC2_155 LR-------------DPSRYLRGGELVCTVGISLQTP-DDCEAFVAALCE
Mb1218c|M.bovis_AF2122/97 AAGSADVFFLIGVTDDEAVRWVDDQARQRAPVAIFVK-HPSDSVVAGAVR
Rv1186c|M.tuberculosis_H37Rv AAGSADVFFLIGVTDDEAVRWVDDQARQRAPVAIFVK-HPSDSVVAGAVR
MMAR_4254|M.marinum_M AAGSADIFFLLGVTDHEALRWIDGQAG--TPVAIFVK-QPSDAVVAKAVR
MUL_0931|M.ulcerans_Agy99 AAGSADIFFLLGVTDHEALRWIDGQAG--TPVAIFVK-QPSDAVVAKAVR
MAV_1330|M.avium_104 AAGSADVFFLLGVGDDEALRWIDTQARDRVPVAVFVK-EPSDALVGTAVA
MLBr_01652|M.leprae_Br4923 -----------------RLKQYSGRLATEAVTAMQER-LPFFADLEASQR
TH_1338|M.thermoresistible__bu -----------------RLKQYSGRLATEAVHTMEER-LPFFADLEASQR
MAB_4202c|M.abscessus_ATCC_199 ---------------DPAQWLSGGELVLTTGIGLPKSSRDRRRYLRSLDE
. . . : :
Mflv_0801|M.gilvum_PYR-GCK AGAAGVCFG--------------------------VGDGHDDVPAALLDA
Mvan_0042|M.vanbaalenii_PYR-1 ARAAGVCFG--------------------------VGDEHDAVPEALLAQ
MSMEG_1068|M.smegmatis_MC2_155 SGAAGLCFG--------------------------TGDAHDAVPDALVAA
Mb1218c|M.bovis_AF2122/97 AGSAVVAVEPRARWERLYHLVNHVLEHHGDRADPTDDSGTDLFGLAQSLA
Rv1186c|M.tuberculosis_H37Rv AGSAVVAVEPRARWERLYHLVNHVLEHHGDRADPTDDSGTDLFGLAQSLA
MMAR_4254|M.marinum_M SGAAVVAVEPQARWERLYRLVNHVLEHHGDRT--TDDSGTDLFGLAQSLA
MUL_0931|M.ulcerans_Agy99 SGAAVVAVEPQARWERLYRLVNHVLEHHGDRT--TDDSGTDLFGLAQSLA
MAV_1330|M.avium_104 AGSAVVAVDPRARWERLYQLVNHVLEHHGDRADAADDSGTDLFGLAQSLA
MLBr_01652|M.leprae_Br4923 ASVTLVVQT-------------------------------AVVNFVQWMQ
TH_1338|M.thermoresistible__bu ASVHLVVQT-------------------------------AVVNFVEWMR
MAB_4202c|M.abscessus_ATCC_199 NNVAALGFG--------------------------TGLTFDEVPPELVST
: .
Mflv_0801|M.gilvum_PYR-GCK CRRAGLPVLVAPPSVPFSVISRFVAESRLGTEIAVARATNALVPELLLSL
Mvan_0042|M.vanbaalenii_PYR-1 CRLHGLPLLVAPPSVPFSVVSRFVAEYWLGSEIAVARATNALVPELLSSL
MSMEG_1068|M.smegmatis_MC2_155 CTRRRLPLLIAPPNVLFATVSRYVADFQVGGELAVARATNTLVPELLASA
Mb1218c|M.bovis_AF2122/97 DRIHGMISIEDAQSHVLAYSASNDEADELRRLSILGRAGPPEHLQWIGQW
Rv1186c|M.tuberculosis_H37Rv DRIHGMISIEDAQSHVLAYSASNDEADELRRLSILGRAGPPEHLQWIGQW
MMAR_4254|M.marinum_M DRIHGMVSIEDAQSHVLAYSASSDEADELRRLSILGRAGPPEHLKWIGQW
MUL_0931|M.ulcerans_Agy99 DRIHGMVSIEDAQSHVLAYSASSDEADELRRLSILGRAGPPEHLKWIGQW
MAV_1330|M.avium_104 ERIHGMVSIENAQSQVLAYSASNDEADELRRLSILGRAGPPEHLEWIGQW
MLBr_01652|M.leprae_Br4923 NPHSDVSYTAQAFELVPQDLAR---RIALRHTVDMVRVTMEFFEEVVPLL
TH_1338|M.thermoresistible__bu DPRSNVTYTMQAFEVVPQDLTR---RIALRQTVEMVRTTMEFFEQVVPLL
MAB_4202c|M.abscessus_ATCC_199 ADELGMPLLEIPLRTPFAAVVKAVSTRIAELEYDAVLRASRAQPRITRAI
: . .
Mflv_0801|M.gilvum_PYR-GCK RRHDSPRRLLDAAGQVLGCYFLLEPEGAAK-GDEA----------AVTVP
Mvan_0042|M.vanbaalenii_PYR-1 RRHDSPRQLLDSAGQLLGCYFLLEPEGGAD-SDDD----------AVTVP
MSMEG_1068|M.smegmatis_MC2_155 RRRESPRQLLDRAGEVLGCYFLLEPGAPADTGGPA----------AHTVT
Mb1218c|M.bovis_AF2122/97 GIFDALRAGREVVRVAERPELGLRPRLAIGIHQPG----------VGALR
Rv1186c|M.tuberculosis_H37Rv GIFDALRPGREVVRVAERPELGLRPRLAIGIHQPG----------VGALR
MMAR_4254|M.marinum_M GIFDALRSGSEVVRVAERPELGLRPRLAIGIHQPP----------VAARR
MUL_0931|M.ulcerans_Agy99 GIFDALRSGSEVVRVAERPELGLRPRLAIGIHQPP----------VAARR
MAV_1330|M.avium_104 GIFDALRSGTQVVRVAERPELGLRPRLAVGIHQPD----------PDARR
MLBr_01652|M.leprae_Br4923 ARSEEQLTALTVGILKYSRDLAFTAATAYADAAEA----------RGTWD
TH_1338|M.thermoresistible__bu ARSEEQVTALTAGILRYSRDLAFAAATAYADAAEA----------RGAWD
MAB_4202c|M.abscessus_ATCC_199 VNGGVQAVTAELGRSLRASVVVLDPGGTVVASHPRNLDVATVNLVRGALS
. : . :
Mflv_0801|M.gilvum_PYR-GCK VPGLGALVWMGRGDRPEPALLDLIAR----FVRAAQGERDIEAALTRERV
Mvan_0042|M.vanbaalenii_PYR-1 VPGLGTLVWVGRGDHPEPALLDLIAR----FVRAAQGERDIEAALARERV
MSMEG_1068|M.smegmatis_MC2_155 IDGLGTLVWVGSGTPPDDAVLDVIVR----FVHAAQGERDIEAALARERV
Mb1218c|M.bovis_AF2122/97 PPVFAGTIWVQQGSQPLADDAEEMLRGAAVLAARIMSRLATQPNTHALRV
Rv1186c|M.tuberculosis_H37Rv PPVFAGTIWVQQGSQPLADDAEEMLRGAAVLAARIMSRLATQPNTHALRV
MMAR_4254|M.marinum_M PPVFAGSIWVQQGSQPLAEDADEVLRGAAVLGARIMTRLASRPSTHARRV
MUL_0931|M.ulcerans_Agy99 PPVFAGSIWVQQGSQPLAEDADEVLRGAAVLGARIMTRLASRPSTHARRV
MAV_1330|M.avium_104 PPVFAGTIWVQQGAQPLADDAEQILRGAAVLAARIMARLAARPSTQARRL
MLBr_01652|M.leprae_Br4923 -------------SRMEASVVDAVVRG--DTGPELLSRAAALNWDTTAPA
TH_1338|M.thermoresistible__bu -------------TRMEANVVDAVVRG--AVGPELQSQAAALNWDATAPA
MAB_4202c|M.abscessus_ATCC_199 PGASAGVQQLDPGITVSQQAIGVGGTSYGVLAVVSKTPLTFVDQVLLGHT
Mflv_0801|M.gilvum_PYR-GCK GQLLSLVER-RMLLP---DALS----------------------------
Mvan_0042|M.vanbaalenii_PYR-1 GQLLSLVER-RMLLP---DALS----------------------------
MSMEG_1068|M.smegmatis_MC2_155 GQLLSLVER-RMLLP---DALN----------------------------
Mb1218c|M.bovis_AF2122/97 QQLLGLAELNATTAP---VDVSTIARELGVAAEGNATLIGFDTAENRDTA
Rv1186c|M.tuberculosis_H37Rv QQLLGLAELNATTAP---VDVSTIARELGVAAEGNATLIGFDTAENRDTA
MMAR_4254|M.marinum_M QQLLGLADPDPTAAP---ADVTAIARELGLSADGNAALIGFDTIEADD--
MUL_0931|M.ulcerans_Agy99 QQLLGLADPDPTAAP---ADVTAIARELGLSADGNAALIGFDTIEADD--
MAV_1330|M.avium_104 QQLLGVTDS-ETLAP---VDLTAIAAELGLAADGCAALVGWAAAEGAS--
MLBr_01652|M.leprae_Br4923 TVVVGTPTPNHDGPNGQVS-------------------------------
TH_1338|M.thermoresistible__bu TVVVGLPRPDR---------------------------------------
MAB_4202c|M.abscessus_ATCC_199 NSLLALDFDKPSRLQEAQRRLNGQALGLLLGDERNLGPVWAQLGQAADAR
::.
Mflv_0801|M.gilvum_PYR-GCK ----QLVQWPGF-AGEIACSAWPAGAGALLSISFPGALIGDAPDLCMMLT
Mvan_0042|M.vanbaalenii_PYR-1 ----QLLEWPGFSAGEIACSAWPAGAGALVSMAFPDALIGDAPDLCLMLT
MSMEG_1068|M.smegmatis_MC2_155 ----QLLDWPGLAAREVACSAWPAGAGALLSMAFPTALVGDAPEVCLVLT
Mb1218c|M.bovis_AF2122/97 VRHVRLVDVMALSASAFRHDAQVAANGSRIYVLLPQTTTGRAVTSWVRGT
Rv1186c|M.tuberculosis_H37Rv VRHVRLVDVMALSASAFRHDAQVAANGSRIYVLLPQTTTGRAVTSWVRGT
MMAR_4254|M.marinum_M -KQSRLADVLALSASAFRRDAQVGTNGSRIYVVLPQTATARSLTSWVRGT
MUL_0931|M.ulcerans_Agy99 -KQSRLADVLALSASAFRRDAQVGTNGSRIYVVLPQTATARSLTSWVRGT
MAV_1330|M.avium_104 -RHTRLTDVIALSASAFRHDAHVAGHGSRTYVLLPQPPN-RSVSSWVRGT
MLBr_01652|M.leprae_Br4923 ------SERASQEVREIAARHGRAALTDVHGTWLVAIISGQLAPTDKFFS
TH_1338|M.thermoresistible__bu ------LEVVSQDVHHVANRHGRTALSDVHGTWLVTIVSGALSPTDRFLT
MAB_4202c|M.abscessus_ATCC_199 --GRIRVLVVDAESQTALVRVLPAVTRDMESAGYPVFAHTGERQLTVVLP
.
Mflv_0801|M.gilvum_PYR-GCK AGAVEMPDDL-------ALPTGHSAPVT-LADLG--SAIAQARIALTLAE
Mvan_0042|M.vanbaalenii_PYR-1 VGALEVTDDL-------ALPTGHSAPVP-LTDIG--SAIAQARIALTLAE
MSMEG_1068|M.smegmatis_MC2_155 DPATSATADLPAIAAELSLPSGHSATAP-LAELG--SAITQARIALDLAR
Mb1218c|M.bovis_AF2122/97 ISALRAELGV-------ALRAAIAGPVAGLAEVN--PARVEVDRVLESAE
Rv1186c|M.tuberculosis_H37Rv ISALRAELGV-------ALRAAIAGPVAGLAEVN--PARVEVDRVLESAE
MMAR_4254|M.marinum_M ISALRSELGV-------ELRATIAAPVAGLDGLA--SARVEVDRVLDSAE
MUL_0931|M.ulcerans_Agy99 ISALRSELGV-------ELRATIAAPVAGLDGLA--SARVEVDRVLDSAE
MAV_1330|M.avium_104 IAALRAELGV-------QLRAVIAAPVPGLSGVA--AARAEVDRVLDSAE
MLBr_01652|M.leprae_Br4923 DLLHAFSDGP----------VVIGPTAPMLTAAY--HSASEAVSGMNAVA
TH_1338|M.thermoresistible__bu DLLDVFADGP----------VVLGPTAPTLSAAY--RSATEAIAGMNAVA
MAB_4202c|M.abscessus_ATCC_199 GAETADFARRLFGDVDARIKKGLRAGLSGAHPVGRLADAVQNARLAASVA
. : .
Mflv_0801|M.gilvum_PYR-GCK HRGGRVGPDQ-LSTLESLLEQLPLAQLAPFRQLLIDPLTELDTSRGTQHV
Mvan_0042|M.vanbaalenii_PYR-1 HRGGRVGPDQ-LSTLESLLEQLPLAQLAPFKQLLIAPLAELDSQRGTQHV
MSMEG_1068|M.smegmatis_MC2_155 QHGGSIGPDQ-LSTFDSLLEGLPLSRLTPFKQQLIDPLAESDHERGTQHV
Mb1218c|M.bovis_AF2122/97 RHPI-LGQVTSLAEARTTVLLDEIVTLVGTDQRLVDPRIRDLGAQDPVLA
Rv1186c|M.tuberculosis_H37Rv RHPI-LGQVTSLAEARTTVLLDEIVTLVGTDQRLVDPRIRDLGAQDPVLA
MMAR_4254|M.marinum_M RHPISIGRVTSLAEARTTVLLDEIVTLVGNDARLIDPRIPDLRAKDPVLA
MUL_0931|M.ulcerans_Agy99 RHPISIGRVTSLAEARTTVLLDEIVTLVGNDARLIDPRIPDLRAKDPVLA
MAV_1330|M.avium_104 RHPLSFGQVTSLAEARTTVLLDEIVTLVGRDERLVDPRIVALHDREPVLA
MLBr_01652|M.leprae_Br4923 GWSGAPRPVQARELLPERALMGDASAIVALHTDVMLPLADAG----PTLI
TH_1338|M.thermoresistible__bu GWSGAPRPVPARELLPERALLGDQTAIAALESEVMRPLADAG----PTLT
MAB_4202c|M.abscessus_ATCC_199 ERGGTPLEFTSLAGSALLSFGASREVLVAVANATLEPIVEHDAVHGSELM
:. : * .
Mflv_0801|M.gilvum_PYR-GCK RTLRTFVACNGSLSETAKELFLHTNTVRHRLGRIQEITGRDPLQHNDLTT
Mvan_0042|M.vanbaalenii_PYR-1 RTLRVFLACNGSLSETAKELFLHTNTVRHRLSRIQEITGRDPLQHNDLTA
MSMEG_1068|M.smegmatis_MC2_155 RTLRVFLATNGSLIDTARELFLHTNTVRHRLARIHEITGRNPLHFEDQAA
Mb1218c|M.bovis_AF2122/97 QTLRAYLDAFGDIGAAARSLQVHPNTVRYRIRRIEQLLSTSLGDPDVRLL
Rv1186c|M.tuberculosis_H37Rv QTLRAYLDAFGDIGAAARSLQVHPNTVRYRIRRIEQLLSTSLGDPDVRLL
MMAR_4254|M.marinum_M DTLRVYLDSFGDIGLAAQSLQVHPNTVRYRIRRVEKLLSTSFADPDVRLV
MUL_0931|M.ulcerans_Agy99 DTLRVYLDSFGDIGLAAQSLQVHPNTVRYRIRRVEKLLSTSFADPDVRLV
MAV_1330|M.avium_104 QTLRTYLDAFGDIAAAAHALRVHPNTVRYRVRRIEKLLSVSLADPEVRLL
MLBr_01652|M.leprae_Br4923 ETLDAYLDCGGAIEACARKLFVHPNTVRYRLKRITDFTGHDPTLPRDAYV
TH_1338|M.thermoresistible__bu ETLDAFLESGGAIEACARRLFVHPNTVRYRLRRIAEITGRDPAVPRDAYV
MAB_4202c|M.abscessus_ATCC_199 TSLRAYLEANGHWESAATVVGVHRHTLRKRIETVQSLLACDLDIARVRAE
:* .:: * * : :* :*:* *: : .: . .
Mflv_0801|M.gilvum_PYR-GCK FVIGMWAADRGE-----------------------------
Mvan_0042|M.vanbaalenii_PYR-1 FVIGLWAADRAERPAGL------------------------
MSMEG_1068|M.smegmatis_MC2_155 FAIGLRASDRHSRMTRRPN----------------------
Mb1218c|M.bovis_AF2122/97 FSLGLRAMERTA-----------------------------
Rv1186c|M.tuberculosis_H37Rv FSLGLRAMERTA-----------------------------
MMAR_4254|M.marinum_M FSLGLRATDRSS-----------------------------
MUL_0931|M.ulcerans_Agy99 FSLGLRATDHSS-----------------------------
MAV_1330|M.avium_104 FALALRAAER-------------------------------
MLBr_01652|M.leprae_Br4923 LRVAATVGKLNYPTHH-------------------------
TH_1338|M.thermoresistible__bu LRVAVSVSRLTRQSYQSSTVNPDLGSTAGWRGQAAGGHRAM
MAB_4202c|M.abscessus_ATCC_199 LLLAMLTGTPGTGN---------------------------
: :. .