For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TITDLPFAVLRLQYRIARTPLQLFESSVISRMDTEAPRRLMFERAVGSIDATVGNLLRDRDLEQSGITRI ERAVELAEAARLDEVADQKKRQAGDELKEKRDQAVAAPQKAREEAKERVQEARAEAEQGKRQAAENVAKR TSQAEEQIDQSAQQRKQNAEKAKQNVQNRSRAAEKAEVKVAEEQLDDAADKRSAAVGARAHADKLEDYSE TEKLKRQAAADKP*
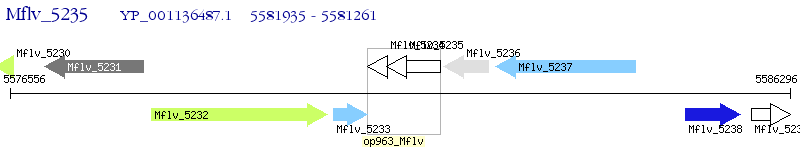
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5235 | - | - | 100% (224) | hypothetical protein Mflv_5235 |
| M. gilvum PYR-GCK | Mflv_5236 | - | 8e-07 | 26.62% (154) | CsbD family protein |
| M. gilvum PYR-GCK | Mflv_4188 | - | 6e-05 | 22.82% (206) | chromosome segregation protein SMC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3777 | - | 2e-06 | 27.01% (174) | Band 7 protein |
| M. marinum M | MMAR_2416 | lgt | 1e-05 | 26.26% (198) | prolipoprotein diacylglyceryl transferases Lgt |
| M. avium 104 | MAV_2471 | - | 2e-05 | 25.55% (137) | DoxX subfamily protein, putative |
| M. smegmatis MC2 155 | MSMEG_1951 | - | 1e-40 | 41.01% (217) | hypothetical protein MSMEG_1951 |
| M. thermoresistible (build 8) | TH_2964 | - | 8e-25 | 39.57% (139) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1486 | - | 8e-05 | 26.88% (160) | cell wall-associated hydrolase |
| M. vanbaalenii PYR-1 | Mvan_1035 | - | 3e-74 | 62.10% (219) | hypothetical protein Mvan_1035 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5235|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1035|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1951|M.smegmatis_MC2_155 --------------------------------------------------
TH_2964|M.thermoresistible__bu --------------------------------------------------
MMAR_2416|M.marinum_M MTATVLAYFPSPPRGVWHLGPLPIRAYAFFIIIGIVMGLMIGDRRFAARG
MAB_3777|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2471|M.avium_104 --------------------------------------------------
MUL_1486|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5235|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1035|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1951|M.smegmatis_MC2_155 --------------------------------------------------
TH_2964|M.thermoresistible__bu --------------------------------------------------
MMAR_2416|M.marinum_M GERGVIYDIALWMVPFGLIGGRLYHLSTDWQTYFGEGGAGLGAALRIWDG
MAB_3777|M.abscessus_ATCC_1997 ---------------------------------------MSSLVIIVIIA
MAV_2471|M.avium_104 --------------------------------------------------
MUL_1486|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5235|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1035|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1951|M.smegmatis_MC2_155 --------------------------------------------------
TH_2964|M.thermoresistible__bu --------------------------------------------------
MMAR_2416|M.marinum_M GLGIWGAVTLGTVGAWIGCRRRGIPLPAFLDAVAPGVVMGQAIGRLGNYF
MAB_3777|M.abscessus_ATCC_1997 IAALVLFVALPLIYVRNYVKVPPNEVAVFTGRGQPKVVRGGARFKMPGIE
MAV_2471|M.avium_104 --------------------------MAFIGQGVNSLLNPKSAAEAAAPA
MUL_1486|M.ulcerans_Agy99 ----------------------------MRRTRRSSASRSAARLVRPAIP
Mflv_5235|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1035|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1951|M.smegmatis_MC2_155 --------------------------------------------------
TH_2964|M.thermoresistible__bu --------------------------------------------------
MMAR_2416|M.marinum_M NQELYGRETTVPWGLEIFYRRDPSGFVDVHSLDGVSTGQVAVVVQPTFLY
MAB_3777|M.abscessus_ATCC_1997 RVDIMSLEPFNVNINLQNALYNNGVPVNVEAVGLVRIGSNDEAVQTAVQR
MAV_2471|M.avium_104 VDGLQALPDPVSGNIPSDPETVAQITAAVQIGG-----------------
MUL_1486|M.ulcerans_Agy99 SVVSVALLVTVPAVANADSRTDSIAALIADVARANQRLDDLSAAVELEQE
Mflv_5235|M.gilvum_PYR-GCK -------------------MTITDLPFAVLR-LQYRIARTPLQLFESSVI
Mvan_1035|M.vanbaalenii_PYR-1 -------------------MSITDIPFAVLR-FQYRLARTPLQLIERRFL
MSMEG_1951|M.smegmatis_MC2_155 -----------------MHMRITDVPFAVLR-LQYQVARLPLQLIEDQVM
TH_2964|M.thermoresistible__bu ----------------------------VLR-LQYQLARFPLQVIGDQFV
MMAR_2416|M.marinum_M ELIWNLLVFAALIYLDRRFTIGHGRLFASYV-ALYCVGRFCVELLRDDTA
MAB_3777|M.abscessus_ATCC_1997 FLTS---------DLSELQRQINEILAGSLRGITATMTVEDLNSNRDSLA
MAV_2471|M.avium_104 ------------------GLLLATGKLPRVASAALALTVLPANLGAHSFW
MUL_1486|M.ulcerans_Agy99 GVNKAMVAVETARDEAAAAEHELEASQQAVKDANAAIAAAQHRFDTFAAA
: .
Mflv_5235|M.gilvum_PYR-GCK SRMD------------------------------------TEAPRRLMFE
Mvan_1035|M.vanbaalenii_PYR-1 TRVD------------------------------------TEAPARLLYE
MSMEG_1951|M.smegmatis_MC2_155 TRLD------------------------------------PETPARLFYE
TH_2964|M.thermoresistible__bu ARLD------------------------------------DESPARLWYE
MMAR_2416|M.marinum_M THIAGIRINSFTSTFVFIGAVVYIILAPKGREDPASVRGTAAAAEEVPEP
MAB_3777|M.abscessus_ATCC_1997 RSVV------------------------------------EEAGGDLARI
MAV_2471|M.avium_104 NESD-------------------------------PVAKAQKRQQFLTDL
MUL_1486|M.ulcerans_Agy99 TYMN-------------------------------------GPSDSYLTA
Mflv_5235|M.gilvum_PYR-GCK RAVGSIDATVGNLLRDRDLEQSGITRIERAVELAEAARL-----------
Mvan_1035|M.vanbaalenii_PYR-1 RAIGAADAAAGSFLRDNELEESGITRIEKASALGEAARL-----------
MSMEG_1951|M.smegmatis_MC2_155 RSFGKLDETVGGALGAVDLRERGSALVERSNALLRAIQL-----------
TH_2964|M.thermoresistible__bu GSLGRLDAVIGGALGDEALRRRGAARMDRSAALGRAARL-----------
MMAR_2416|M.marinum_M EPAGEVATAAAPAGDTEGDEPTGLAETAKAAEVAEPETVGSAVAGDESTA
MAB_3777|M.abscessus_ATCC_1997 GMEVDVLKIAGISDRNGYLESLGQRRIAEVRRDATVGTA-----------
MAV_2471|M.avium_104 SLLGGLLIASADTAGKPSLGWRGRRAAERLSERVSSAWPGSD--------
MUL_1486|M.ulcerans_Agy99 TSPDEIIAAATAAKTLAASAQTVMANLERARTRQVNKES-----------
.
Mflv_5235|M.gilvum_PYR-GCK -----DEVADQKKRQAGDELKEKRDQAVAAPQKAREEAKERVQEARAEAE
Mvan_1035|M.vanbaalenii_PYR-1 -----DEVAAQQKEQAADELQRKRDKAATAPQKAGQEARQRVQNARQKAD
MSMEG_1951|M.smegmatis_MC2_155 -----EAEATRNTREADAELREAQAEAAEDLKDARADREQNVKEARSTAL
TH_2964|M.thermoresistible__bu -----DDTATERVQAADAELRHTREQAEQRQHAAQATTQQQVREARETAE
MMAR_2416|M.marinum_M DTEASDAVESEVGEAAEAEAPEAEAEEAEVGEVEVAEAVEGEVEEAEEAE
MAB_3777|M.abscessus_ATCC_1997 -----EAERDAQIQSAQARQAGAIAQAEADTAIATATQKRDVELARLRAQ
MAV_2471|M.avium_104 ----DSGFEADLSELGERIAHGLQIGAERGRELASTAAERGAPYAEAALE
MUL_1486|M.ulcerans_Agy99 ----ASRLAEQKADKAAEDAKTSQDAAVTALTDTQRKFDQQREEVNRLAA
. . .
Mflv_5235|M.gilvum_PYR-GCK QGKRQAAENVAKRTSQAEEQIDQSAQQRKQNAEKAKQNVQNRSRAAEKAE
Mvan_1035|M.vanbaalenii_PYR-1 KRTQDVAQSVASRTAQAKREVDEAAGRKVEAAEKAKRSMQNRSRAAEKVV
MSMEG_1951|M.smegmatis_MC2_155 QRKQAAEQARQKRTQTAKAKTDEVAAKRKEVAQTAKQRKEEAINTVEQAK
TH_2964|M.thermoresistible__bu NRKRDAAVTAQR--GTAAAKRQAXXXXRGGTRRRPRRRN------LRRSR
MMAR_2416|M.marinum_M PGEVEVAEAVEGEVEAAAEAEEAEEAEEAEEAEEAEEAEPGEIEVAEAVE
MAB_3777|M.abscessus_ATCC_1997 TEAENAQADQAGPLAQARAEKDVGIAREQAEAARVQARTEVEQRRTEQAQ
MAV_2471|M.avium_104 RGRELASTAAERGAPYAEAALERGRELASTAAERGRPLAKKARKRGEELA
MUL_1486|M.ulcerans_Agy99 ERDEAEAKLQAARLVAWSSAGAEGPPPGAMWDPGARPGNGRRWDGWDPTL
.
Mflv_5235|M.gilvum_PYR-GCK -----VKVAEEQLDDAADKR-SAAVGARAHADKLEDYSETEKLKRQAAAD
Mvan_1035|M.vanbaalenii_PYR-1 -----TSVAEKELDDAASKR-SAAAGARAHADRLADFSEAEKDKRQAEKS
MSMEG_1951|M.smegmatis_MC2_155 -----TKNARAKLADAQETR-VEAAEKIVEADQIEEWAEAEKAKRS----
TH_2964|M.thermoresistible__bu -----PRHGSRR--------------------------------------
MMAR_2416|M.marinum_M GEV--EAAAEAEEAEVGEVE-AVDEAAEAEAPEVGEIEAAEAVEGEVEVA
MAB_3777|M.abscessus_ATCC_1997 AALQADVIAPAEARRQADIA-IAEGARQAAILKAQSDAEAERQKGAAEAD
MAV_2471|M.avium_104 DEAAERAAYLAQKARKRSEE-LAEEAAERGKPLAKKARKRSEKLAKKARK
MUL_1486|M.ulcerans_Agy99 PQIPSANIPGDPIAVVNQVLGISATSAQVTANMGQNFLQQLGILKPTDTG
Mflv_5235|M.gilvum_PYR-GCK KP------------------------------------------------
Mvan_1035|M.vanbaalenii_PYR-1 E-------------------------------------------------
MSMEG_1951|M.smegmatis_MC2_155 --------------------------------------------------
TH_2964|M.thermoresistible__bu --------------------------------------------------
MMAR_2416|M.marinum_M AEAEEAEAGEDE---PKEAEEVAEAEGSEAEDKTVQAIDGGQTGVDDAGE
MAB_3777|M.abscessus_ATCC_1997 ARKAAADAFRVE---QQAAADGAKAKLIAEADGTKAKLLADAEGIKAKLL
MAV_2471|M.avium_104 RSEKLAGTARVR---GEELAETARQRGSLLADTARERVGGQVGTGRRKLS
MUL_1486|M.ulcerans_Agy99 ITNAATGATRIPRVYGRQASEYVIRRGMAQIGVPYSWGGGNAAGPSKGID
Mflv_5235|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1035|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1951|M.smegmatis_MC2_155 --------------------------------------------------
TH_2964|M.thermoresistible__bu --------------------------------------------------
MMAR_2416|M.marinum_M ADAEPDDDSTAAEPEVVVEQADIEEIEPQEPEAEAVAAAEATDADEGDGE
MAB_3777|M.abscessus_ATCC_1997 AEADGKKEVAAALNAYTPEAARLLTLPDILNSVVQATEAASKPIGEIENL
MAV_2471|M.avium_104 W-------------------------------------------------
MUL_1486|M.ulcerans_Agy99 SGAGITGFDCSGLVLYSFAGVGIKLPHYSGSQYNLGRKIPTSQMRRGDVI
Mflv_5235|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1035|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1951|M.smegmatis_MC2_155 --------------------------------------------------
TH_2964|M.thermoresistible__bu --------------------------------------------------
MMAR_2416|M.marinum_M PAEPASDDLDAVAAEDADVDVDATATEAEESGDESPEPAADSEEPEADES
MAB_3777|M.abscessus_ATCC_1997 SIIGGAGDAQDALGTLFGVSPQAVAKVIESLRVSGIDVPDLLKRTQQADA
MAV_2471|M.avium_104 --------------------------------------------------
MUL_1486|M.ulcerans_Agy99 FYGPGGSQHVTLYLGNGQMLEAPDIGLKVRVAPVRTSGMTPFVVRYIEY-
Mflv_5235|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1035|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1951|M.smegmatis_MC2_155 --------------------------------------------------
TH_2964|M.thermoresistible__bu --------------------------------------------------
MMAR_2416|M.marinum_M APEDSAEDTSSGESDTQSELEQSQTIEQGSDQTSAGDGEGPPPSAAVDGD
MAB_3777|M.abscessus_ATCC_1997 PASPAPAPAVHPEGWVQE--------------------------------
MAV_2471|M.avium_104 --------------------------------------------------
MUL_1486|M.ulcerans_Agy99 --------------------------------------------------
Mflv_5235|M.gilvum_PYR-GCK ---------------------
Mvan_1035|M.vanbaalenii_PYR-1 ---------------------
MSMEG_1951|M.smegmatis_MC2_155 ---------------------
TH_2964|M.thermoresistible__bu ---------------------
MMAR_2416|M.marinum_M TGPTRQGWRGRLRNRMRRSGR
MAB_3777|M.abscessus_ATCC_1997 ---------------------
MAV_2471|M.avium_104 ---------------------
MUL_1486|M.ulcerans_Agy99 ---------------------