For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSIDTAYMRDALTRRLLRRGVSGGTITLPAVPAMVDEYVSMCNKIFAALGRRFNADELSHLRSLLAKELA NAFDGSNRSNIVISYDAPIGNVLNYHIQPQWTSLADTYDNWVATRKPPLFGTHPDARVTTLAAEIADPTT ARVLDIGAGTGRNALALAKKGHPVDAVELTPKFADILREEAYKHLLGVRVIDRDIFETLDDLQKDYSLII LSEVTPDFRSPEQIRAVFELAAQCLAPGGRLVFNVFMPRPGFMSDDAVNELADSARQLAQQVYSAIFFGD EIDTAGAGLPLRLMSNDSVYEFEKENLPPGAWPHTGWYAEWVSGQDIFDVPREQSPVEMRWLVYQKTG
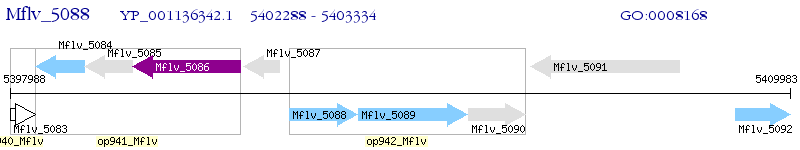
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5088 | - | - | 100% (348) | methyltransferase type 12 |
| M. gilvum PYR-GCK | Mflv_3090 | - | 6e-08 | 32.26% (124) | methyltransferase type 11 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2446 | - | 1e-116 | 57.31% (342) | hypothetical protein Mb2446 |
| M. tuberculosis H37Rv | Rv2423 | - | 1e-116 | 57.31% (342) | hypothetical protein Rv2423 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3750 | - | 1e-115 | 57.35% (340) | hypothetical protein MMAR_3750 |
| M. avium 104 | MAV_2748 | - | 1e-105 | 51.73% (346) | hypothetical protein MAV_2748 |
| M. smegmatis MC2 155 | MSMEG_5176 | - | 3e-06 | 31.47% (143) | methyltransferase type 11 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1267 | - | 1e-163 | 78.74% (348) | methyltransferase type 12 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5088|M.gilvum_PYR-GCK ------MSIDTA-YMRDALTRRLLRRGVSGGTITLPAVPAMVDEYVSMCN
Mvan_1267|M.vanbaalenii_PYR-1 ----MLMSIDTA-YMREAMARRLLRRGVSSGKITLPAVPGLVDEYVSVCN
Mb2446|M.bovis_AF2122/97 MDNLPIESAESTRLAKAAMTRRFYTRSVVKGEITLPAVPSMIDEYVTMCA
Rv2423|M.tuberculosis_H37Rv MDNLPIESAESTRLAKAAMTRRFYTRSVVKGEITLPAVPSMIDEYVTMCA
MMAR_3750|M.marinum_M ---MPLSSVDSQQSLKNSMTRRFYSRSEVTGKITLPAVPAMIDEYVKMCD
MAV_2748|M.avium_104 MQDPPNRSFDPS-LLREAVIRRIYRRTSAEGTIRVPAVPAMIDEYLQLCG
MSMEG_5176|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_5088|M.gilvum_PYR-GCK KIFAALGRRFNADELSHLRSLLAKELANAFDGSNRSNIVISYDAPIGNVL
Mvan_1267|M.vanbaalenii_PYR-1 KIFGALGRKFSGDELDHLRSLLAKELANAYAGSNRSNIVITYDAPVGTVL
Mb2446|M.bovis_AF2122/97 GLFAGVGRKFSDEELAHLRAVLQGQLAEAYAASQRSTIVISYNAPMGPTL
Rv2423|M.tuberculosis_H37Rv GLFAGVGRKFSDEELAHLRAVLQGQLAEAYAASQRSTIVISYNAPMGPTL
MMAR_3750|M.marinum_M GMFADVGRKFSEEELAHLRSVLEGQLAQAYSASPRSSIIISFNAPVGPTL
MAV_2748|M.avium_104 NVCATLSVWYAPEQFAQLRSALEVELAKAFKASPRSDVLISYHAPFGTGV
MSMEG_5176|M.smegmatis_MC2_155 -------------------------------------------------M
:
Mflv_5088|M.gilvum_PYR-GCK NYHIQPQWTSLADTYDNWVATRKPPLFGTHPDARVTTLAAEIADPTTARV
Mvan_1267|M.vanbaalenii_PYR-1 NYFIQPQVSTLEESYDSWVASRKPPLFGINPDARVTALSTEITSPETARV
Mb2446|M.bovis_AF2122/97 HYQVRAQWRTVAQEYENWIATREPPLFGTEPDARVWALANEAADPTTHRV
Rv2423|M.tuberculosis_H37Rv HYQVRAQWRTVAQEYENWIATREPPLFGTEPDARVWALANEAADPTTHRV
MMAR_3750|M.marinum_M NYEVNARWWTVEAAYENWIDTRKPPLFGTEPDARVWALANEAADPSTHRV
MAV_2748|M.avium_104 NFHVQADWRTIEADYEQWVAVRPPPLFGTEPDARVLTLAAEADDPATYRV
MSMEG_5176|M.smegmatis_MC2_155 PPTDAAQPEPVDNPFFAWLWKT----LSTHETESVRRMRAENLAGLSGRV
. .: : *: :. . * : * : **
Mflv_5088|M.gilvum_PYR-GCK LDIGAGTGRNALALAKKGHPVDAVELTPKFADILREEAYKHLLGVRVIDR
Mvan_1267|M.vanbaalenii_PYR-1 LDIGAGTGRNALALARRGHPVDAVELTPKFAEILRTEAFKHLLNIRVIDR
Mb2446|M.bovis_AF2122/97 LEIGAGTGRNALALARRGHPVDVVEMTPKFADIIRSDAERDSLDVRVIMR
Rv2423|M.tuberculosis_H37Rv LEIGAGTGRNALALARRGHPVDVVEMTPKFADIIRSDAERDSLDVRVIMR
MMAR_3750|M.marinum_M LEIGAGTGRNALALARRGHPVDVVELTPKFAEMIRADAEKEGLDVRVIVR
MAV_2748|M.avium_104 LDVGAGTGRNALALARRGHPVDAVEMTAKFAEAMRADAERESLGVNVIQS
MSMEG_5176|M.smegmatis_MC2_155 LEVGAGTGTNFEFYPSTVDEVVAIEPERRLADVARQAAAKAPVPVSVTG-
*::***** * . . * .:* ::*: * * : : : *
Mflv_5088|M.gilvum_PYR-GCK DIFETLDDLQKDYSLIILSEVTPDFRSPEQIRAVFELAAQCLAPGGRLVF
Mvan_1267|M.vanbaalenii_PYR-1 DVFETMDDLQKDYQLIVLSEVVSDFRNVAQLRAIFELAAGCLAPGGRLVF
Mb2446|M.bovis_AF2122/97 DVFSTMDDLRQDYQLMVLSEVVPDFRTTQQLRNLFELAAQCLAPGARLVF
Rv2423|M.tuberculosis_H37Rv DVFSTMDDLRQDYQLMVLSEVVPDFRTTQQLRNLFELAAQCLAPGARLVF
MMAR_3750|M.marinum_M DVFTTIADLRRDYQLIVLSEVVSDFRTTQQLRELFELAAQCLAPGARLVF
MAV_2748|M.avium_104 DVFTAMENTRDRYQLMVLSEVVPDFRTAHELRGMFELAAECLAPGGRLVF
MSMEG_5176|M.smegmatis_MC2_155 DTIETFASAQP-FDAVVCSLVLCSVDDPE---QVVGQLYSLLRPGGELRY
* : :: . : :. :: * * .. :. * **..* :
Mflv_5088|M.gilvum_PYR-GCK NVFMPRPGFMSDDAVNELADSARQLAQQVYSAIFFGDEIDTAGAGLPLRL
Mvan_1267|M.vanbaalenii_PYR-1 NTFMPRPGYLSDDAVRELADAARELGQQVYTTIFTGEEIDIASAGLSLRL
Mb2446|M.bovis_AF2122/97 NAFLANGDYAPD-------QAAREFGQQMYTGMCTRAEMSAAAAGLPLEL
Rv2423|M.tuberculosis_H37Rv NAFLANGDYAPD-------QAAREFGQQMYTGMCTRAEMSAAAAGLPLEL
MMAR_3750|M.marinum_M NAFLACGGYTPD-------QAAREFGQQAYTTIFTRHELSVAAAGLPVEL
MAV_2748|M.avium_104 NTFLARDGYVPD-------AAAVQFSQQCNSMVFTRDEVMGAAGGLPLAL
MSMEG_5176|M.smegmatis_MC2_155 LEHVAGTGFQAR--------LQRLADATVWPKLFGGCHTHRHTEQVIASS
.:. .: . . : . :
Mflv_5088|M.gilvum_PYR-GCK MSNDSVYEFEKENLPPGAWPHTGWYAEWVSGQDIFDVPREQSPVEMRWLV
Mvan_1267|M.vanbaalenii_PYR-1 MTNDSVYEYEKENLPAGHWPPTGWYAEWVSGQDIFDVPREQSPVDMRWLV
Mb2446|M.bovis_AF2122/97 VADDSVYDYEKTHLPPGAWPPTSWYADWIRGLDVFTTNVESCPIEMRWLV
Rv2423|M.tuberculosis_H37Rv VADDSVYDYEKTHLPPGAWPPTSWYADWIRGLDVFTTNVESCPIEMRWLV
MMAR_3750|M.marinum_M IADDSVYDYEKAHLPADAWPPTSWYADWVSGLDVFPVAREMSPIELRWLV
MAV_2748|M.avium_104 VADDSAYEYEKTHSPAESWPPTGWFEGWAGGLDVFDVERDDSPIELRWIV
MSMEG_5176|M.smegmatis_MC2_155 GFG--VRGRRREQVLPAWVPVP---------TTEFAIGRAVRP-------
. . .: : . * . * *
Mflv_5088|M.gilvum_PYR-GCK YQKTG--
Mvan_1267|M.vanbaalenii_PYR-1 YQKTG--
Mb2446|M.bovis_AF2122/97 FQRRR--
Rv2423|M.tuberculosis_H37Rv FQRRR--
MMAR_3750|M.marinum_M FRKTH--
MAV_2748|M.avium_104 FAKTRAG
MSMEG_5176|M.smegmatis_MC2_155 -------