For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGEQPGDLADPTPFEVGKHQVDQAVVLTVSGEVDMLSSPMLAEAVQTALAAKPAALIVDLSKVGFLASAG MTVLVTTQAELQPPTKFAVVADGSATSRPIKLMGIDSVLSLYPSLNDALSALSGA
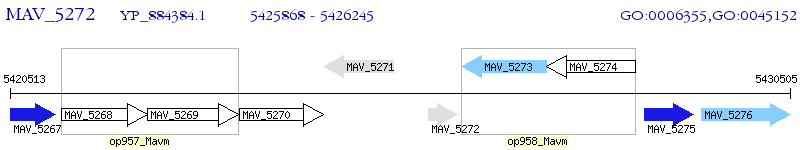
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5272 | - | - | 100% (125) | anti-anti-sigma factor |
| M. avium 104 | MAV_0435 | - | 2e-21 | 46.53% (101) | anti-anti-sigma factor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3712c | - | 1e-14 | 46.05% (76) | hypothetical protein Mb3712c |
| M. gilvum PYR-GCK | Mflv_2021 | - | 7e-25 | 50.47% (107) | anti-sigma-factor antagonist |
| M. tuberculosis H37Rv | Rv3687c | rsfB | 2e-20 | 39.32% (117) | anti-anti-sigma factor RSFB (anti-sigma factor antagonist) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0214 | - | 1e-36 | 61.79% (123) | hypothetical protein MMAR_0214 |
| M. smegmatis MC2 155 | MSMEG_6127 | - | 2e-25 | 49.56% (113) | anti-anti-sigma factor |
| M. thermoresistible (build 8) | TH_1832 | - | 2e-24 | 50.50% (101) | ANTI-ANTI-SIGMA FACTOR RSFB (ANTI-SIGMA FACTOR ANTAGONIST) |
| M. ulcerans Agy99 | MUL_3602 | - | 4e-07 | 31.48% (108) | anti sigma factor antagonist |
| M. vanbaalenii PYR-1 | Mvan_3003 | - | 4e-25 | 46.67% (120) | anti-sigma-factor antagonist |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3712c|M.bovis_AF2122/97 --------------------------------------------------
Rv3687c|M.tuberculosis_H37Rv -------------MSAPDSITVTVADHNGVAVLSIGGEIDLITAAALEEA
TH_1832|M.thermoresistible__bu -----------------------VEHRDDSAVLRVQGVVDLATGDALQQA
MAV_5272|M.avium_104 MGEQP------GDLADPTPFEVGKHQVDQAVVLTVSGEVDMLSSPMLAEA
MMAR_0214|M.marinum_M MVEQS------GDPTPPTAFGIEKSRVGEAVVVTVSGELDMLTAPRLAEM
MSMEG_6127|M.smegmatis_MC2_155 MTSQ-----------DPANCTVEERRVGDITVVAVTGTVDMLTAPKLEDA
Mvan_3003|M.vanbaalenii_PYR-1 MDEQAAEGTNVGGAASSSTCVVTERWVERTAVVAVSGVVDMLTSPQLESA
Mflv_2021|M.gilvum_PYR-GCK ------------------MQQFTEREVDGVVIVAAVGAIDMLTAPQLQEA
MUL_3602|M.ulcerans_Agy99 ----------------MDLLAVEHEVRQDTVVVRAKGDVDSSTVEQLVVH
Mb3712c|M.bovis_AF2122/97 ----MADNPTA----LVIDLSAVEFLGSVGLKILAATSEK-IGQSVKFGV
Rv3687c|M.tuberculosis_H37Rv IGEVVADNPTA----LVIDLSAVEFLGSVGLKILAATSEK-IGQSVKFGV
TH_1832|M.thermoresistible__bu IDDLIATRPSA----LVVDLSAVDFLASVGLQILVATHER-LSPSARFAV
MAV_5272|M.avium_104 VQTALAAKPAA----LIVDLSKVGFLASAGMTVLVTTQAE-LQPPTKFAV
MMAR_0214|M.marinum_M LRTSLIEAPAT----LIVDLSKVDFLASAGMTVLVSAKAQ-AVAPTRFVV
MSMEG_6127|M.smegmatis_MC2_155 IGSAAKSEPSA----VVVDLSAVDFLASAGMGVLVAAHGE-LAPAVRLVV
Mvan_3003|M.vanbaalenii_PYR-1 IDTALSQQPAA----VVIDFTDVEFLASAGMGVLVAAHDK-AGSDVKISV
Mflv_2021|M.gilvum_PYR-GCK VAAAARRRPSG----LVVDLTSVDFLGSAGMQVLMETRKG-LDGDTRFAV
MUL_3602|M.ulcerans_Agy99 LAAAMQLAATHPARLVVIDLQPVTFFGSAGLNAVLECHEQGMAAGTSVRL
.: :::*: * *:.*.*: : . . :
Mb3712c|M.bovis_AF2122/97 VARGSVTRRPIHLMGLDKTFRLFSTLHDALTGVRGGRIDR-------
Rv3687c|M.tuberculosis_H37Rv VARGSVTRRPIHLMGLDKTFRLFSTLHDALTGVRGGRIDR-------
TH_1832|M.thermoresistible__bu VADGPATSRPIQLTRLDEVIALYPTLDEALSGVRTGERAE-------
MAV_5272|M.avium_104 VADGSATSRPIKLMGIDSVLSLYPSLNDALSALSGA-----------
MMAR_0214|M.marinum_M VADGPATSRPIRLMGIDAMLPLYDALDDALNDISDS-----------
MSMEG_6127|M.smegmatis_MC2_155 VADGPATSRPLKLVGIADVVDLFATLDEALSSLKT------------
Mvan_3003|M.vanbaalenii_PYR-1 VAEGPATSRPLKLVGIADIVALYPNLEEALAARDT------------
Mflv_2021|M.gilvum_PYR-GCK VADGPATSRPLKITGITDYVDLYPSLDVALADLAG------------
MUL_3602|M.ulcerans_Agy99 VADHGQLLAPIQVTELDRIFEIYPTLAGALRPGKPGEHDLEDVERPR
** *::: : . :: * **