For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSQDPANCTVEERRVGDITVVAVTGTVDMLTAPKLEDAIGSAAKSEPSAVVVDLSAVDFLASAGMGVLV AAHGELAPAVRLVVVADGPATSRPLKLVGIADVVDLFATLDEALSSLKT
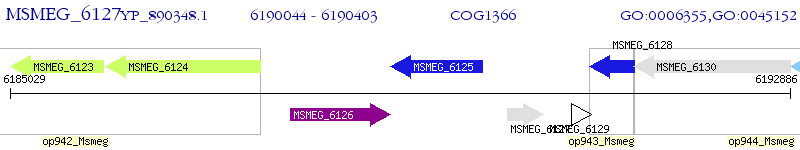
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6127 | - | - | 100% (119) | anti-anti-sigma factor |
| M. smegmatis MC2 155 | MSMEG_5551 | - | 5e-08 | 32.48% (117) | stas domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3712c | - | 8e-12 | 42.25% (71) | hypothetical protein Mb3712c |
| M. gilvum PYR-GCK | Mflv_3284 | - | 5e-33 | 61.06% (113) | anti-sigma-factor antagonist |
| M. tuberculosis H37Rv | Rv3687c | rsfB | 4e-19 | 38.05% (113) | anti-anti-sigma factor RSFB (anti-sigma factor antagonist) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0214 | - | 8e-26 | 49.11% (112) | hypothetical protein MMAR_0214 |
| M. avium 104 | MAV_5272 | - | 1e-25 | 49.56% (113) | anti-anti-sigma factor |
| M. thermoresistible (build 8) | TH_0498 | - | 5e-35 | 65.14% (109) | anti-anti-sigma factor |
| M. ulcerans Agy99 | MUL_4427 | - | 5e-07 | 25.42% (118) | hypothetical protein MUL_4427 |
| M. vanbaalenii PYR-1 | Mvan_3003 | - | 1e-31 | 59.29% (113) | anti-sigma-factor antagonist |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3712c|M.bovis_AF2122/97 --------------------------------------------------
Rv3687c|M.tuberculosis_H37Rv MSAP-------------DSITVTVADHNGVAVLSIGGEIDLITAAALEEA
Mflv_3284|M.gilvum_PYR-GCK MDEQAAEGTAAGGAP-SSTCVVTQRWVDTTAVVAVTGVVDMLTSPQLETA
Mvan_3003|M.vanbaalenii_PYR-1 MDEQAAEGTNVGGAASSSTCVVTERWVERTAVVAVSGVVDMLTSPQLESA
TH_0498|M.thermoresistible__bu --------------------VIDERWDADIAVVSATGVVDMLSAPQLDEA
MSMEG_6127|M.smegmatis_MC2_155 MTSQDPAN-----------CTVEERRVGDITVVAVTGTVDMLTAPKLEDA
MMAR_0214|M.marinum_M MVEQSGDPTPP------TAFGIEKSRVGEAVVVTVSGELDMLTAPRLAEM
MAV_5272|M.avium_104 MGEQPGDLADP------TPFEVGKHQVDQAVVLTVSGEVDMLSSPMLAEA
MUL_4427|M.ulcerans_Agy99 MNATAKSP---------NALAIDTRSEDSLVVLSVEGAIDSTNCAALRDA
Mb3712c|M.bovis_AF2122/97 ----MADNPTALVIDLSAVEFLGSVGLKILAATSEKIGQ--SVKFGVVAR
Rv3687c|M.tuberculosis_H37Rv IGEVVADNPTALVIDLSAVEFLGSVGLKILAATSEKIGQ--SVKFGVVAR
Mflv_3284|M.gilvum_PYR-GCK IDDALNQKPGAVVIDFTEVEFLASAGMGVLVAAHDKAGS--EVKISVVAD
Mvan_3003|M.vanbaalenii_PYR-1 IDTALSQQPAAVVIDFTDVEFLASAGMGVLVAAHDKAGS--DVKISVVAE
TH_0498|M.thermoresistible__bu IRTALGKKPRALVVDFTAVDFLASAGMGVLVAIHDEVSP--AVNLTIVAD
MSMEG_6127|M.smegmatis_MC2_155 IGSAAKSEPSAVVVDLSAVDFLASAGMGVLVAAHGELAP--AVRLVVVAD
MMAR_0214|M.marinum_M LRTSLIEAPATLIVDLSKVDFLASAGMTVLVSAKAQAVA--PTRFVVVAD
MAV_5272|M.avium_104 VQTALAAKPAALIVDLSKVGFLASAGMTVLVTTQAELQP--PTKFAVVAD
MUL_4427|M.ulcerans_Agy99 IIKATLDEPSAVVVNVSALQVPDEASWSIFVSARWQVDTPQHVPILLVCA
* :::::.: : . ... ::.: : . : :*.
Mb3712c|M.bovis_AF2122/97 GSVTRRPIHLMGLDKTFRLFSTLHDALTGVRGGRIDR-------------
Rv3687c|M.tuberculosis_H37Rv GSVTRRPIHLMGLDKTFRLFSTLHDALTGVRGGRIDR-------------
Mflv_3284|M.gilvum_PYR-GCK GPATSRPLKLVGIADIVALYSNLDEALAARDT------------------
Mvan_3003|M.vanbaalenii_PYR-1 GPATSRPLKLVGIADIVALYPNLEEALAARDT------------------
TH_0498|M.thermoresistible__bu GPATSRPLKLLGIADLVPLHATLDEALAAVAT------------------
MSMEG_6127|M.smegmatis_MC2_155 GPATSRPLKLVGIADVVDLFATLDEALSSLKT------------------
MMAR_0214|M.marinum_M GPATSRPIRLMGIDAMLPLYDALDDALNDISDS-----------------
MAV_5272|M.avium_104 GSATSRPIKLMGIDSVLSLYPSLNDALSALSGA-----------------
MUL_4427|M.ulcerans_Agy99 SRAGRELITRTGVTRFMPVYPTEKRAIKAVGRLARRKVRYAQVQLPANLG
. . . : *: . :. . *:
Mb3712c|M.bovis_AF2122/97 --------------------------------------------------
Rv3687c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3284|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3003|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0498|M.thermoresistible__bu --------------------------------------------------
MSMEG_6127|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0214|M.marinum_M --------------------------------------------------
MAV_5272|M.avium_104 --------------------------------------------------
MUL_4427|M.ulcerans_Agy99 SLRESRKLVREWLTSWSKPGLIPVALVIVNVFVENVLEHTGSDPVIKVQC
Mb3712c|M.bovis_AF2122/97 --------------------------------------------------
Rv3687c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3284|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3003|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0498|M.thermoresistible__bu --------------------------------------------------
MSMEG_6127|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0214|M.marinum_M --------------------------------------------------
MAV_5272|M.avium_104 --------------------------------------------------
MUL_4427|M.ulcerans_Agy99 DGTAATIAVSDGSNAPAIRLQTPPKGIDVSGLAIVEALCRAWGSTPTATG
Mb3712c|M.bovis_AF2122/97 -------------
Rv3687c|M.tuberculosis_H37Rv -------------
Mflv_3284|M.gilvum_PYR-GCK -------------
Mvan_3003|M.vanbaalenii_PYR-1 -------------
TH_0498|M.thermoresistible__bu -------------
MSMEG_6127|M.smegmatis_MC2_155 -------------
MMAR_0214|M.marinum_M -------------
MAV_5272|M.avium_104 -------------
MUL_4427|M.ulcerans_Agy99 KTVWAIIGPENQL