For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
CRRATGSAITTFGRNNFGFNNFGLNAVEDRTVHGDKPHFYGIARVIRAASIPIILIWVAIAAFLNVSVPQ LEEVGKLRSVSMSPDDAPAVISMQRIGEVFEEYESNSSVMIVLEGQEPLGDDTRAYYSELIADLEADTKH VEHIQDLWSDPLTAAGAQSADGMSTYFQVYLAGNQGEALANESVRAVQELVAESQPPDGVAAYVTGASAL AAEQEEAGHSSMRFVEALTFLVITIMLVLFFRSIVTTLLILVMVGLSLMTVRGAVAFLGFHDIIGLSTFA TSLLVTLAIAIAVDYAIFLIGRYQEAKGTGATSEEAYYTMFGGTAHVIVGSGLTIAGATFCLSFTRLPYF QSLGVPLALGMLVLIAVAMTFGPAVVTVASKFGLLAPKRDMRVRGWRKVGAATVRWPGAILVASVAVSLI GLLALPGYQTSYNDRKYLPADIQSNIGYAAAERHFSPARLNPEVLMVETDHDLRNPADFIVIEKIAKALF AVEGIGRVQTITRPDGKPIESTSIPYIMSRQNTTQQLNEKYMLDRMDDMRAQADSLQTTINVMERMQALM TQMSAITTSMVGKTRTMTADITQLRDHIADLDDFVRPIRNYFYWEPHCYNIPVCHSMRSLFDLIDGTNLL TENTQELLPDLERMAAIMPELIALMPSQIESMKSQRDNMLTQYQTQNSQLEAGAEMSVDSGAMGDAFNDA WNADTFYLPPEAFENKDFQSGIEQFISPNGHAVRFIIAHEDDPLSPAGIEKIDGLKTAAKEAIKGTPLEG ARVYLGGTAATFKDMADGTRYDLLIAGIAAIGLIFIIMLILTRAVIAAAVIVGTVVLSLGASFGLSVLVW QHFVGIELHWMVLPMAVIILLAVGADYNLLVVSRLKEEIQAGVGTGLIRTMGGSGSVVTAAGMVFALTMM TMAVSDLTIIGQVGTTIGMGLIFDTLVIRSFMTPSIAALLGRWFWWPQKIRTRPVPVPWGAGASGGGDGD GDGDGDDGGGGAPRARASAQ*
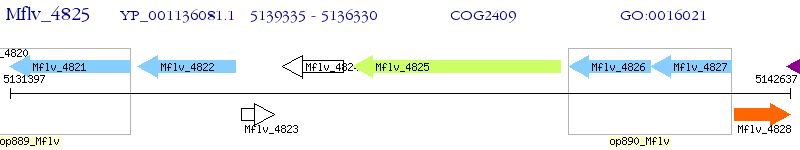
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4825 | - | - | 100% (1001) | transport protein |
| M. gilvum PYR-GCK | Mflv_3435 | - | 0.0 | 68.18% (949) | transport protein |
| M. gilvum PYR-GCK | Mflv_3029 | - | 0.0 | 61.28% (935) | transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0695c | mmpL5 | 0.0 | 66.06% (934) | transmembrane transport protein MmpL5 |
| M. tuberculosis H37Rv | Rv0676c | mmpL5 | 0.0 | 65.95% (934) | transmembrane transport protein MmpL5 |
| M. leprae Br4923 | MLBr_02378 | - | 0.0 | 56.56% (930) | hypothetical protein MLBr_02378 |
| M. abscessus ATCC 19977 | MAB_4382c | - | 0.0 | 60.82% (929) | putative membrane protein, MmpL |
| M. marinum M | MMAR_1005 | mmpL5 | 0.0 | 65.12% (929) | transmembrane transport protein, MmpL5 |
| M. avium 104 | MAV_2510 | - | 0.0 | 64.48% (929) | MmpL4 protein |
| M. smegmatis MC2 155 | MSMEG_0225 | - | 0.0 | 66.17% (940) | MmpL4 protein |
| M. thermoresistible (build 8) | TH_3260 | - | 0.0 | 66.14% (942) | PUTATIVE MmpL4 protein |
| M. ulcerans Agy99 | MUL_0757 | mmpL5 | 0.0 | 65.02% (929) | transmembrane transport protein, MmpL5 |
| M. vanbaalenii PYR-1 | Mvan_1614 | - | 0.0 | 89.85% (946) | transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4825|M.gilvum_PYR-GCK MCRRATGSAITTFGRNNFGFNNFGLNAVEDRTVHGDKPHFYGIARVIRAA
Mvan_1614|M.vanbaalenii_PYR-1 ---------MSRLGDD----DQPSMSSLSGESPR--KPHFYGIARVIRRA
TH_3260|M.thermoresistible__bu --------MTANPGHVR---TDEAPTDRLDIKVSTHGPSF--ISTWIRRL
MSMEG_0225|M.smegmatis_MC2_155 ---------MSTPDAP----TEAIPSAHGKYTNHG------GIAKLIRTL
MAB_4382c|M.abscessus_ATCC_199 ---------MSKPMNDP--TTDAFPAARHAARPW--------LPRMIRVL
Mb0695c|M.bovis_AF2122/97 --------MIVQRTAAP---TGSVPPDRHAARPF--------IPRMIRTF
Rv0676c|M.tuberculosis_H37Rv --------MIVQRTAAP---TGSVPPDRHAARPF--------IPRMIRTF
MMAR_1005|M.marinum_M --------MTATTTDTP---TDVVPPAEHPPRPF--------IPRMIRTF
MUL_0757|M.ulcerans_Agy99 --------MTATTTDTP---TDVVPPAEHPPRPF--------IPRMIRTF
MAV_2510|M.avium_104 --------MSTTFDDTR---TDAIPVVKEPVREK--------IPRFIRAF
MLBr_02378|M.leprae_Br4923 --------MTVKCANDL---------DTHTKPPF--------VARMIHRF
:. *:
Mflv_4825|M.gilvum_PYR-GCK SIPIILIWVAIAAFLNVSVPQLEEVGKLRSVSMSPDDAPAVISMQRIGEV
Mvan_1614|M.vanbaalenii_PYR-1 SIPIILSWIALAAFLNISVPQLEEVGKQRSVSMSPDDAPAVISMEHIGEV
TH_3260|M.thermoresistible__bu SLPIIVGWIALIALLNITVPQLEVVGEMRAVSMSPDEAPSMVAMKRVGEL
MSMEG_0225|M.smegmatis_MC2_155 AVPIILVWVALIAVLNTIVPQLEEVGEMRSVSMSPDDAPSMIAMKRVGEV
MAB_4382c|M.abscessus_ATCC_199 AVPIVLGWIAVTIALNTVVPQLEEVGHMRAVSMSPNDAPAMIATKRVGEV
Mb0695c|M.bovis_AF2122/97 AVPIILGWLVTIAVLNVTVPQLETVGQIQAVSMSPDAAPSMISMKHIGKV
Rv0676c|M.tuberculosis_H37Rv AVPIILGWLVTIAVLNVTVPQLETVGQIQAVSMSPDAAPSMISMKHIGKV
MMAR_1005|M.marinum_M AVPLILGWVLLIAVLNSVVPQLEVVGQMQAVSMSPDAAPSMISMKHIGKV
MUL_0757|M.ulcerans_Agy99 AVPLILGWVLLIAVLNSVVPQLEVVGQMQAVSMSPDAAPSMISMKHIGKV
MAV_2510|M.avium_104 AVPIIIGWIALIAVLNVVVPQLDEVGKMRSVEMTPDDAQSVVATKRMGAV
MLBr_02378|M.leprae_Br4923 AVPIILVWLAIAVTVSVFIPSLEDVGQERSVSLSPKDAPSYIAMQKMGQV
::*::: *: :. :*.*: **. ::*.::*. * : :: :::* :
Mflv_4825|M.gilvum_PYR-GCK FEEYESNSSVMIVLEGQEPLGDDTRAYYSELIADLEADTKHVEHIQDLWS
Mvan_1614|M.vanbaalenii_PYR-1 FEEYESNSSVMIVLEGEQPLGDDTRRYYADLISGLEADTTHVEHVQDLWS
TH_3260|M.thermoresistible__bu FDEGSSDSQVMIVLEGEEPLGADARAFYDELVKRLEADDAHVENVQDLWS
MSMEG_0225|M.smegmatis_MC2_155 FEEFKSNSSVMIVLEGEQPLGDEAHKYYDEIVDKLEADKTHIEHVQDFWG
MAB_4382c|M.abscessus_ATCC_199 FEEFKTSSSVMIVLEGEQSLGADAHRFYDQMVRDLRADTTHVQHVQDFWA
Mb0695c|M.bovis_AF2122/97 FEEGDSDSAAMIVLEGQRPLGDAAHAFYDQMIGRLQADTTHVQSLQDFWG
Rv0676c|M.tuberculosis_H37Rv FEEGDSDSAAMIVLEGQRPLGDAAHAFYDQMIGRLQADTTHVQSLQDFWG
MMAR_1005|M.marinum_M FREGDSDSAAMIVLEGQEPLGAAAHKFYDKMITKLRADTKHVQSVQDFWG
MUL_0757|M.ulcerans_Agy99 FREGDSDSAAMIVLEGQEPLGAAAHKFYDKMITKLRADTKHVQSVQDFWG
MAV_2510|M.avium_104 FNEYKSNSSVMIVLEGQQPLGADAHAYYDEIVRRLNADTKHVEHVQDMWS
MLBr_02378|M.leprae_Br4923 FNEGNSDSVIMIVLEGDKPLGDDAHRFYDGLIRKLRAD-KHVQSVQDFWG
* * .:.* ******:..** :: :* :: *.** *:: :**:*.
Mflv_4825|M.gilvum_PYR-GCK DPLTAAGAQSADGMSTYFQVYLAGNQGEALANESVRAVQELVA---ESQP
Mvan_1614|M.vanbaalenii_PYR-1 DPLTAAGAQSADGKSTYFQVYLAGNQGEALANESVRAVQELVA---ESQP
TH_3260|M.thermoresistible__bu DPLTATGVQSPDGKSAYVQVKLAGNQGEALANESVETVREMIATVSEEMA
MSMEG_0225|M.smegmatis_MC2_155 DPLTASGAQSSDGLASYVQVYTAGNQGEALANESVKAVQDIVD---SVQA
MAB_4382c|M.abscessus_ATCC_199 DALTAAAAQSSDGKAAYVQVYIAGDQGEALANESVEAVRHITA---AVPP
Mb0695c|M.bovis_AF2122/97 DPLTATGAQSSDGKAAYVQVKLAGNQGESLANESVEAVKTIVE---RLAP
Rv0676c|M.tuberculosis_H37Rv DPLTATGAQSSDGKAAYVQVKLAGNQGESLANESVEAVKTIVE---RLAP
MMAR_1005|M.marinum_M DPLTATGAQSGDGKAAYVQVKLAGNQGESLANESVEAVKTIVD---SLQA
MUL_0757|M.ulcerans_Agy99 DPLTATGAQSGDGKAAYVQVKLAGNQGESLANESVEAVKTIVD---SLQA
MAV_2510|M.avium_104 DPLTGAGAQSNDGKASYVQVYLAGNQGEALANESVESVQNIVK---SVQA
MLBr_02378|M.leprae_Br4923 DPLTAPGAQSNDGKAAYVQLSLAGNQGELLAQESIDAVRKIVV---QTPA
*.**....** ** ::*.*: **:*** **:**: :*: : .
Mflv_4825|M.gilvum_PYR-GCK PDGVAAYVTGASALAAEQEEAGHSSMRFVEALTFLVITIMLVLFFRSIVT
Mvan_1614|M.vanbaalenii_PYR-1 PAGVKAFVTGASALAAEQEEAGHTSMRFVEALTFLVITIMLVLFFRSIVT
TH_3260|M.thermoresistible__bu PEGVQAFVTGGAAMSADQQEAGDSSLRVIEALTFLVITTMLLFVYRSVIT
MSMEG_0225|M.smegmatis_MC2_155 PPGVTAYVTGPAAMSADQQIAGDRSMRMIEALTFTVIIIMLLMVYRSIIT
MAB_4382c|M.abscessus_ATCC_199 SPGVRVYVTGPATTSADQEIVANDSMELIELVTFGVIMVMLLLVYRSVIT
Mb0695c|M.bovis_AF2122/97 PPGVKVYVTGSAALVADQQQAGDRSLQVIEAVTFTVIIVMLLLVYRSIIT
Rv0676c|M.tuberculosis_H37Rv PPGVKVYVTGSAALVADQQQAGDRSLQVIEAVTFTVIIVMLLLVYRSIIT
MMAR_1005|M.marinum_M PPGVKVYVTGSAALVADQQLAGDRSLQLIEMVTFTVIIVMLLLVYRSIIT
MUL_0757|M.ulcerans_Agy99 PPGVKVYVTGSAALVADQQLAGDRSLQLIEMVTFTVIIVMLLLVYRSIIT
MAV_2510|M.avium_104 PNGVKAYVTGPAALSADQHTAGDRSLQLITAATFTVIIGMLLLVYRSVIT
MLBr_02378|M.leprae_Br4923 PPGITAWVTGASALIADMHHSGDKSMIRITATSVIVILVTLLLVYRSFIT
. *: .:*** :: *: . .. *: : :. ** *::.:**.:*
Mflv_4825|M.gilvum_PYR-GCK TLLILVMVGLSLMTVRGAVAFLGFHDIIGLSTFATSLLVTLAIAIAVDYA
Mvan_1614|M.vanbaalenii_PYR-1 TLLILVMVGLSLMTVRGAVAFLGFHDIIGLSTFATSLLVTLAIAIAVDYA
TH_3260|M.thermoresistible__bu VLIALFMVLIGLLTTRGVVAFLGYHNIIGLSTFATSLLVTLAIAIAVDYA
MSMEG_0225|M.smegmatis_MC2_155 VILSLVMVVFSLAAARGVIAFLGYYEIIGLSTFATNLLVTLAIAAATDYA
MAB_4382c|M.abscessus_ATCC_199 MVIVVAMVMLELASARGLVALLSYHNAFGLTSFATSLVVTLAIAAATDYA
Mb0695c|M.bovis_AF2122/97 SAIMLTMVVLGLLATRGGVAFLGFHRIIGLSTFATNLLVVLAIAAATDYA
Rv0676c|M.tuberculosis_H37Rv SAIMLTMVVLGLLATRGGVAFLGFHRIIGLSTFATNLLVVLAIAAATDYA
MMAR_1005|M.marinum_M AAIMLTMVVLGLLATRGGVAFLGYHRIIGLSTFATNLLVVLAIAAATDYA
MUL_0757|M.ulcerans_Agy99 AAIMLTMVVLGLLATRGGVAFLGYHRIIGLSTFATNLLVVLAIAAATDYA
MAV_2510|M.avium_104 VLLTLVMVVLELSAARGMVAFLGYYKIIGLSTFATNLLVTLAIAAATDYA
MLBr_02378|M.leprae_Br4923 VILLLFTVGIESAVARGVVALLGHTGLIGLSTFAVNLLTSLAIAAGTDYG
: : * : .** :*:*.. :**::**..*:. **** ..**.
Mflv_4825|M.gilvum_PYR-GCK IFLIGRYQEAKGTGATSEEAYYTMFGGTAHVIVGSGLTIAGATFCLSFTR
Mvan_1614|M.vanbaalenii_PYR-1 IFLVGRYQEAKGKGATAEEAYFTMFGGNAHVIVGSGLTIAGATFCLSFTR
TH_3260|M.thermoresistible__bu IFLIGRYQEARSVGADRATAYHTMFGGTAHVILASGLTIAGATFCLSFTR
MSMEG_0225|M.smegmatis_MC2_155 IFLIGRYQEARSVGESREQAYYTMFHSTAHVVLGSGMTIAGATLCLHFTR
MAB_4382c|M.abscessus_ATCC_199 IFLIGRYQEARRAGEDRESAYYTMFHGTAHVVLASGLTIAGATFCLHFTR
Mb0695c|M.bovis_AF2122/97 IFLIGRYQEARGLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTR
Rv0676c|M.tuberculosis_H37Rv IFLIGRYQEARGLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTR
MMAR_1005|M.marinum_M IFLIGRYQEARGLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTR
MUL_0757|M.ulcerans_Agy99 IFLIGRYQEARGLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTR
MAV_2510|M.avium_104 IFLIGRYQEARAVGESREDAYYTMYKGTAHVVAGSGMTIAGATFCLHFTN
MLBr_02378|M.leprae_Br4923 IFITGRYQEARQANENKEAAFYTMYRGTFHVILGSGLTISGATFCLSFAR
**: ******: . *:.**: .. **: .**:**:***:** *:.
Mflv_4825|M.gilvum_PYR-GCK LPYFQSLGVPLALGMLVLIAVAMTFGPAVVTVASKFG-LLAPKRDMRVRG
Mvan_1614|M.vanbaalenii_PYR-1 LPYFQSLGVPLALGMLVLIAVAMTFGPAMVTIASKFG-LLEPKRAMRVRG
TH_3260|M.thermoresistible__bu LPYFQSLGPPLAIGMIVLIVTSLTMGPAIITVATRFG-LLEPKRAARVRF
MSMEG_0225|M.smegmatis_MC2_155 MPYFQSLGIPLAIGMTVVVLTALAVGPAIITVASRFGKTLEPKRAMRTRG
MAB_4382c|M.abscessus_ATCC_199 LPYFKTTGIPLSIGMAIVVAMALTLGPALISVASRFGNVLEPKGAGKARG
Mb0695c|M.bovis_AF2122/97 LPYFQTLGVPLAIGMVIVVAAALTLGPAIIAVTSRFGKLLEPKRMARVRG
Rv0676c|M.tuberculosis_H37Rv LPYFQTLGVPLAIGMVIVVAAALTLGPAIIAVTSRFGKLLEPKRMARVRG
MMAR_1005|M.marinum_M LPYFQTLGVPLAIGMVIVVATALTLGPALIAVMSRFGKLLEPKRMARVRG
MUL_0757|M.ulcerans_Agy99 LPYFQTLGVPLAIGMVIVVATALTLGPALIAVMSRFGKLLEPKRMARVRG
MAV_2510|M.avium_104 LPYFQTLGIPLAIGMVVVVAAALTLGPAVISVASRFRQTLEPRRTQRIRG
MLBr_02378|M.leprae_Br4923 MPYFQTLGVPCAVGMLIAVAVALTLGPAVLTVGSRFG-LFEPKRLIKVRG
:***:: * * ::** : : :::.***:::: ::* : *: : *
Mflv_4825|M.gilvum_PYR-GCK WRKVGAATVRWPGAILVASVAVSLIGLLALPGYQTSYNDRKYLPADIQSN
Mvan_1614|M.vanbaalenii_PYR-1 WRKIGAVTVRWPGAILVASVAVSLIGLLALPGYQTSYNDRNYLPDDIQSN
TH_3260|M.thermoresistible__bu WRKVGASVVRWPGPIMVTTVAVTLVGLLALPGYRTNYNDRNYLPASFPSN
MSMEG_0225|M.smegmatis_MC2_155 WRKVGAAVVRWPGPILVAAVGLSMIGLLALPGYRTNYNDRDYQPADLPAN
MAB_4382c|M.abscessus_ATCC_199 WRRVGAATVRWPGAILVAAVTMALVGLLTLPGYHTTYNDRIYLPGQVPAN
Mb0695c|M.bovis_AF2122/97 WRKVGAAIVRWPGPILVGAVALALVGLLTLPGYRTNYNDRNYLPADLPAN
Rv0676c|M.tuberculosis_H37Rv WRKVGAAIVRWPGPILVGAVALALVGLLTLPGYRTNYNDRNYLPADLPAN
MMAR_1005|M.marinum_M WRKIGAAIVRWPGPILIGAVALALVGLLTLPGYKTNYNDRNYLPADLPAN
MUL_0757|M.ulcerans_Agy99 WRKIGAAIVRWPGPILIGAVALALVGLLTLPGYKTNYNDRNYLPADLPAN
MAV_2510|M.avium_104 WRKVGTVVVRWPGPILVLTIGVALVGLLTLPGYRTNYNDRNYLPTDLPAN
MLBr_02378|M.leprae_Br4923 WRRIGTVVVRWPLPILITTCAIAMVGLLALPGYRTNYKDRAYLPASIPAN
**::*: **** .*:: : ::::***:****:*.*:** * * .. :*
Mflv_4825|M.gilvum_PYR-GCK IGYAAAERHFSPARLNPEVLMVETDHDLRNPADFIVIEKIAKALFAVEGI
Mvan_1614|M.vanbaalenii_PYR-1 VGYAAAERHFSPARLNPEVLMVETDHDLRNPADFIVIEKIAKALFTVEGI
TH_3260|M.thermoresistible__bu AGYEAAERHFDPARLNPELLLIESDHDLRNPADFLVIEKIAKAVFQVEGI
MSMEG_0225|M.smegmatis_MC2_155 SGYAAADRHFSQARMNPELLMIESDHDLRNSADFLVIERIAKRVVGVPGV
MAB_4382c|M.abscessus_ATCC_199 VGYAAADRHFSKAKMNPDLLLVESDHDLRNPADFLVIDKIAKALARVRGI
Mb0695c|M.bovis_AF2122/97 EGYAAAERHFSQARMNPEVLMVESDHDMRNSADFLVINKIAKAIFAVEGI
Rv0676c|M.tuberculosis_H37Rv EGYAAAERHFSQARMNPEVLMVESDHDMRNSADFLVINKIAKAIFAVEGI
MMAR_1005|M.marinum_M EGYSAAERHFSQARMNPEVLMVESDHDMRNSSDFLVINKIAKAIFGVEGI
MUL_0757|M.ulcerans_Agy99 EGYSAAERHFSQARMNPEVLMVESDHDMRNSSDFLVINKIAKAIFGVEGI
MAV_2510|M.avium_104 EGYAAADRHFSQARMNPEVLMIESDHDLRNSADFLVIDKIAKTVFRVPGI
MLBr_02378|M.leprae_Br4923 QGFAAADRHFPQARMKPEILMIESDHDMRNPADFLILDKLARGIFRVPGI
*: **:*** *:::*::*::*:***:**.:**::::::*: : * *:
Mflv_4825|M.gilvum_PYR-GCK GRVQTITRPDGKPIESTSIPYIMSRQNTTQQLNEKYMLDRMDDMRAQADS
Mvan_1614|M.vanbaalenii_PYR-1 GRVQTITRPDGTPIESTSIPYIMSRQSTTQQLNEKYMLDRMDDMQVQAES
TH_3260|M.thermoresistible__bu SRVQGITRPEGTPIEHSSIPFMISQQGTLQILNQQYMQDRMADMLVQADE
MSMEG_0225|M.smegmatis_MC2_155 SRVQAITRPQGTPIEHTSIPFTISMQGTTQTMNQKYMQDRMADMLVQADQ
MAB_4382c|M.abscessus_ATCC_199 AQVQTITRPDGKPIKHTSIPFNMSTANTAQTLTNDYSQANFANLLKQADA
Mb0695c|M.bovis_AF2122/97 SRVQAITRPDGKPIEHTSIPFLISMQGTSQKLTEKYNQDLTARMLEQVND
Rv0676c|M.tuberculosis_H37Rv SRVQAITRPDGKPIEHTSIPFLISMQGTSQKLTEKYNQDLTARMLEQVND
MMAR_1005|M.marinum_M SRVQAITRPDGKPIEHTSIPFLISMQGTSQKLTEKYNQDLTTSMLQQVND
MUL_0757|M.ulcerans_Agy99 SRVQAITRPDGKPIEHTSIPFLISMQGTSQKLTEKYNQDLTTSMLQQVND
MAV_2510|M.avium_104 GRVQAITRPQGTPIEHTSIPFQISMQGVTQQMNQKYQQDQMADMLHQADM
MLBr_02378|M.leprae_Br4923 SRVQAITRPDGTAMDHTSIPFQISMQNAGQVQTMKYQKDRMNDLLRQAEN
.:** ****:*..:. :***: :* .. * . .* : *.:
Mflv_4825|M.gilvum_PYR-GCK LQTTINVMERMQALMTQMSAITTSMVGKTRTMTADITQLRDHIADLDDFV
Mvan_1614|M.vanbaalenii_PYR-1 LQTTIDTMEKMQALMSQMSEITNSMVAKTKTMAVDITELRDHIADLDDFV
TH_3260|M.thermoresistible__bu MQLMIDTMDKMIALMEEMVETTDNMVRDMDEMAAHVAEMRDSISEFDDFF
MSMEG_0225|M.smegmatis_MC2_155 MQVTVDTMTKMIALMEQMRDTMNSMVGKMHGMVDDIKELRDNISDFDDFF
MAB_4382c|M.abscessus_ATCC_199 MQVTIDSMEKMQAITQQMAEVTASMSANMKNTAQDINQLRDNMSNFDDQF
Mb0695c|M.bovis_AF2122/97 IQSNIDQMERMHSLTQQMADVTHEMVIQMTGMVVDVEELRNHIADFDDFF
Rv0676c|M.tuberculosis_H37Rv IQSNIDQMERMHSLTQQMADVTHEMVIQMTGMVVDVEELRNHIADFDDFF
MMAR_1005|M.marinum_M IQTNIDQMERMHDLTQQMADVTHQMVIQMKGMVVDVEELRNHIADFDDFF
MUL_0757|M.ulcerans_Agy99 IQTNIDQMERMHDLTQQMADVTHQMVIQMKGMVVDVEELRNHIADFDDFF
MAV_2510|M.avium_104 MQTTIDSMEKMQSITVQMSNDMHVMVQKMHDMTIDINDLRNKMADFEDFF
MLBr_02378|M.leprae_Br4923 MAETIASMRRMHQLMALLTENTHHILNDTVEMQKTTSKLRDEIANFDDFW
: : * :* : : : . .:*: :::::*
Mflv_4825|M.gilvum_PYR-GCK RPIRNYFYWEPHCYNIPVCHSMRSLFDLIDGTNLLTENTQELLPDLERMA
Mvan_1614|M.vanbaalenii_PYR-1 RPIRNYFYWEPHCYNIPVCHSMRSLFDLIDGTNLLTDNVQDLVPDLERMA
TH_3260|M.thermoresistible__bu RPIRNYFYWEPHCYNIPVCWAMRSVFDTLDGINTMTDDIQNMLPHMHRLN
MSMEG_0225|M.smegmatis_MC2_155 RPIRNYLYWEPHCYNIPMCYSLRSIFDTMDGIDTMTADFEQIVPDMDRMN
MAB_4382c|M.abscessus_ATCC_199 RPIRNYFYWEKHCFDIPMCWALRSVFDALDGISLMSDDFDTLVPDIQRMA
Mb0695c|M.bovis_AF2122/97 RPIRSYFYWEKHCYDIPVCWSLRSVFDTLDGIDVMTEDINNLLPLMQRLD
Rv0676c|M.tuberculosis_H37Rv RPIRSYFYWEKHCYDIPVCWSLRSVFDTLDGIDVMTEDINNLLPLMQRLD
MMAR_1005|M.marinum_M RPIRSYFYWEKHCFDIPVCWSLRSVFDTLDGVDLMTEDINNLLPLMERLD
MUL_0757|M.ulcerans_Agy99 RPIRSYFYWEKHCFDIPVCWSLRSVFDTLDGVDLMTEDINNLLPLMERLD
MAV_2510|M.avium_104 RPMRSYFYWEKHCYDIPVCWSLRSVFDALDGIDTMTDDIQSLLPIMDHLD
MLBr_02378|M.leprae_Br4923 RPIRSYFYWERHCFNIPICWSFRSIFDALDGVDQIDERLNSIVGDIKNMD
**:*.*:*** **::**:* ::**:** :** . : : :: :..:
Mflv_4825|M.gilvum_PYR-GCK AIMPELIALMPSQIESMKSQRDNMLTQYQTQNSQLEAGAEMSVDSGAMGD
Mvan_1614|M.vanbaalenii_PYR-1 AIMPELIALMPQQIETMKSQREMMLTQFQTQKGQLEAAAEMQVDAGAMGD
TH_3260|M.thermoresistible__bu DLMPQMLTLLPTTVATMKTMRSMLLTMQATNAGLQDQMAEMQENANAMGE
MSMEG_0225|M.smegmatis_MC2_155 ALIPVMIANMEPMITTMKTMKTMMLTMQATNAGLQDQMAAMQENSTAMGE
MAB_4382c|M.abscessus_ATCC_199 GLMPQMVVLMPPMIASMKEQKQIMLSQYQVQKAQQDQTMAMQANASAMGQ
Mb0695c|M.bovis_AF2122/97 TLMPQLTAMMPEMIQTMKSMKAQMLSMHSTQEGLQDQMAAMQEDSAAMGE
Rv0676c|M.tuberculosis_H37Rv TLMPQLTAMMPEMIQTMKSMKAQMLSMHSTQEGLQDQMAAMQEDSAAMGE
MMAR_1005|M.marinum_M TLMPQMTAMMPEMIQTMKSMKAQMLSMHSTQQGLQDQMAAMQEDSSAMGE
MUL_0757|M.ulcerans_Agy99 TLMPQMTAMMPEMIQTMKSMKAQMLSMHSTQQGLQDQMAAMQEDSSAMGE
MAV_2510|M.avium_104 TLMPQMVALMPSMIENMKAMKTTMLTMYSTQKGLQDQQNEAQKNSNAMGK
MLBr_02378|M.leprae_Br4923 LLMPQMLEQFPPMIESMESMRTIMLTMHSTMSGIFDQMNELSDNANTMGK
::* : : : .*: : :*: . . : . :: :**.
Mflv_4825|M.gilvum_PYR-GCK AFNDAWNADTFYLPPEAFENKDFQSGIEQFISPNGHAVRFIIAHEDDPLS
Mvan_1614|M.vanbaalenii_PYR-1 AFNDAWNADSFYLPPEAFENADFQSGIEQFISPNGHAVRFIIAHEGDPLS
TH_3260|M.thermoresistible__bu AFDQARNDDSFYLPPEAFDNPDFQRGLEQFLSPDGHAVRFIISHQGDPMT
MSMEG_0225|M.smegmatis_MC2_155 AFDKAKNDDSFYLPPETFDNPDFKRGMKMFLSPDGHAVRFIISHEGDPMS
MAB_4382c|M.abscessus_ATCC_199 AFDDAKNDDSFYLPPEAFETADFKSGIKLFVSPNGHAVRFTIFHQSDPLT
Mb0695c|M.bovis_AF2122/97 AFDASRNDDSFYLPPEVFDNPDFQRGLEQFLSPDGHAVRFIISHEGDPMS
Rv0676c|M.tuberculosis_H37Rv AFDASRNDDSFYLPPEVFDNPDFQRGLEQFLSPDGHAVRFIISHEGDPMS
MMAR_1005|M.marinum_M AFDSSRNDDSFYLPPEVFNNPDFKRGLEQFLSPDGHAVRFIISHEGDPMS
MUL_0757|M.ulcerans_Agy99 AFDSSRNDDSFYLPPEVFNNPDFKRGLEQFLSPDGHAVRFIISHEGDPMS
MAV_2510|M.avium_104 AFDASKNDDSFYLPPETFNNKEFKKGMKNFISPDGHAVRFIISHDGDPMT
MLBr_02378|M.leprae_Br4923 AFDTAKNDDSFYLPPEVFKNTDFKRAMKSFLSSDGHAARFIILHRGDPAS
**: : * *:******.*.. :*: .:: *:*.:***.** * * .** :
Mflv_4825|M.gilvum_PYR-GCK PAGIEKIDGLKTAAKEAIKGTPLEGARVYLGGTAATFKDMADGTRYDLLI
Mvan_1614|M.vanbaalenii_PYR-1 PAGVEKIDKLKTAAKEAVKGTPLEGSKIYLGGTAATFKDMADGTRYDLLI
TH_3260|M.thermoresistible__bu PEGLAKIEPIKKAAVEAIKGTPLEGSRVYLGGTAAVFKDMSDGTKYDLII
MSMEG_0225|M.smegmatis_MC2_155 PEGIQKIDDIKLAAKEAIKGTPLEGSKIYLGGTAATFKDMQEGANYDLII
MAB_4382c|M.abscessus_ATCC_199 EEGTSRVEPLKTAAEDAIKGTPLEGSSIYVGGSAALYKDMQEGADYDLLI
Mb0695c|M.bovis_AF2122/97 QAGIARIAKIKTAAKEAIKGTPLEGSAIYLGGTAAMFKDLSDGNTYDLMI
Rv0676c|M.tuberculosis_H37Rv QAGIARIAKIKTAAKEAIKGTPLEGSAIYLGGTAAMFKDLSDGNTYDLMI
MMAR_1005|M.marinum_M QAGINHIDKIKTAAKEAIKGTPLEGSTIYLGGTAAMFKDLSDGNTFDLMI
MUL_0757|M.ulcerans_Agy99 QAGINHIDKIKTAAKEAIKGTPLEGSTIYLGGTVAMFKDLSDGNTFDLMI
MAV_2510|M.avium_104 QEGISHIGAIKKAAYEALKGTPLEGSKIYLAGTASIYKDLSDGNNYDLLI
MLBr_02378|M.leprae_Br4923 VAGIASINAIRTAAEEALKGTPLEDTKIYLAGTAAVFKDIDEGANWDLVI
* : :: ** :*:******.: :*:.*:.: :**: :* :**:*
Mflv_4825|M.gilvum_PYR-GCK AGIAAIGLIFIIMLILTRAVIAAAVIVGTVVLSLGASFGLSVLVWQHFVG
Mvan_1614|M.vanbaalenii_PYR-1 AGIAAVGLIFIIMLILTRAIVAAAVIVGTVVLSLGASFGLSVLVWQHLVG
TH_3260|M.thermoresistible__bu AAIAAVCLIFIIMLILTRAVVAAAVIVGTVTLSLAASFGVSVLIWQYIVG
MSMEG_0225|M.smegmatis_MC2_155 AGIAALCLIFIIMLIITRAVVASAVIVGTVVISLGSAFGLSVLIWQHLVG
MAB_4382c|M.abscessus_ATCC_199 AAVAALILIFLIMVLLTRAVVAAAVIVGTVVLSLGASFGLSVLLWQHIIG
Mb0695c|M.bovis_AF2122/97 AGISALCLIFIIMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILG
Rv0676c|M.tuberculosis_H37Rv AGISALCLIFIIMLITTRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILG
MMAR_1005|M.marinum_M AGISALCLIFIIMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILG
MUL_0757|M.ulcerans_Agy99 AGISALCLIFIIMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHVLG
MAV_2510|M.avium_104 AGISSLCLIFIIMLIITRGVVASAVIVGTVLLSLGASFGLSVLIWQHLIG
MLBr_02378|M.leprae_Br4923 AGISSLCLIFIIMLIITRAFVAAAVIVGTVALSLGASFGLSVLLWQHILG
*.:::: ***:**:: **..:*:******* :**.::**:***:**:.:*
Mflv_4825|M.gilvum_PYR-GCK IELHWMVLPMAVIILLAVGADYNLLVVSRLKEEIQAGVGTGLIRTMGGSG
Mvan_1614|M.vanbaalenii_PYR-1 IELHWMVLPMAVIILLAVGADYNLLVVSRLKEEVGAGLHTGLIRTMGGSG
TH_3260|M.thermoresistible__bu LEMHWMVFPMALIILLAVGADYNLLVVARLREEIPAGLNTGMIRAMGGSG
MSMEG_0225|M.smegmatis_MC2_155 LELHWMVLAMAVIVLLAVGADYNLLLVSRIKEEIHAGLDTGMIRAMGGSG
MAB_4382c|M.abscessus_ATCC_199 TPLHWMVLPMAVIILLAVGADYNLLLVSRMKEELHAGLHTGTIRSMAGTG
Mb0695c|M.bovis_AF2122/97 IELHWLVLAMAVIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSG
Rv0676c|M.tuberculosis_H37Rv IELHWLVLAMAVIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSG
MMAR_1005|M.marinum_M IELHWLVLAMAVIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSG
MUL_0757|M.ulcerans_Agy99 IELHWLVLAMAVIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSG
MAV_2510|M.avium_104 IELHWMVLAMSVIILLAVGADYNLLLVARFKEEIHAGLNTGIIRSMGGTG
MLBr_02378|M.leprae_Br4923 IELHYLVLAMSVIVLLAVGSDYNLLLVSRFKQEIQAGLKTGIIRSMGGTG
:*::*:.*::*:*****:*****:*:*:::*: **: ** **:*.*:*
Mflv_4825|M.gilvum_PYR-GCK SVVTAAGMVFALTMMTMAVSDLTIIGQVGTTIGMGLIFDTLVIRSFMTPS
Mvan_1614|M.vanbaalenii_PYR-1 SVVTAAGMVFALTMMTMAVSDLTIIGQVGTTIGMGLIFDTLVIRSFMTPS
TH_3260|M.thermoresistible__bu SVVTAAGLVFGLTMMVMVVSDLTVMGQVGTTIGLGLLFDTFVVRSFMMPS
MSMEG_0225|M.smegmatis_MC2_155 SVVTAAGLVFAFTMMSMAVSELAIIGQVGTTIGLGLLFDTLVIRSFMTPS
MAB_4382c|M.abscessus_ATCC_199 SVVTSAGLVFAFTMMGMMVSSFTVIGQIGTTIGLGLLFDTLVVRSFMTPS
Mb0695c|M.bovis_AF2122/97 SVVTAAGLVFAFTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPS
Rv0676c|M.tuberculosis_H37Rv SVVTAAGLVFAFTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPS
MMAR_1005|M.marinum_M SVVTAAGLVFAFTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPS
MUL_0757|M.ulcerans_Agy99 SVVTAAGLVFAFTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPS
MAV_2510|M.avium_104 SVVTSAGLVFAFTMMTMAVSELTVIGQVGTTIGLGLLFDTLIVRSLMTPS
MLBr_02378|M.leprae_Br4923 KVVTNAGLVFAFTMASMVVSDLRVIGQVGTTIGLGLLFDTLIVRSFMMPS
.*** **:**.:** : **.: ::.*:*****:**:***:::**:* **
Mflv_4825|M.gilvum_PYR-GCK IAALLGRWFWWPQKIRTRPVPVPWGAGASGGGDGDGDGDGDDGGGGAPRA
Mvan_1614|M.vanbaalenii_PYR-1 IAALLGRWFWWPQQIRTRPAPSPWPAPPARVADDTGR-------------
TH_3260|M.thermoresistible__bu IATLLGRWFWWPQFVRKRPVPQPWPSVPPRGPATTDRVSV----------
MSMEG_0225|M.smegmatis_MC2_155 IAALLGKWFWWPQMVRRRPKPSPWPKPIQREPADAVN-------------
MAB_4382c|M.abscessus_ATCC_199 IASLLGRWFWWPMVVRPRPRPQPWPQPLHAGAAEAVRA------------
Mb0695c|M.bovis_AF2122/97 IAALLGKWFWWPQVVRQRPVPQPWPSPASARTFALV--------------
Rv0676c|M.tuberculosis_H37Rv IAALLGKWFWWPQVVRQRPIPQPWPSPASARTFALV--------------
MMAR_1005|M.marinum_M IAALLGKWFWWPQVVRQRPAPQPWPTPAPTP--ATTSV------------
MUL_0757|M.ulcerans_Agy99 IAALLGKWFWWPQVVRQRPAPQPWPTPAPTPPPATTSV------------
MAV_2510|M.avium_104 IAALLGKWFWWPQRVRQRPVPSPWPKPAAQEREVLADA------------
MLBr_02378|M.leprae_Br4923 IAALLGRWFWWPQQGRTRPLLTVAAPVGLPPDRSLVYSD-----------
**:***:***** * **
Mflv_4825|M.gilvum_PYR-GCK RASAQ
Mvan_1614|M.vanbaalenii_PYR-1 -----
TH_3260|M.thermoresistible__bu -----
MSMEG_0225|M.smegmatis_MC2_155 -----
MAB_4382c|M.abscessus_ATCC_199 -----
Mb0695c|M.bovis_AF2122/97 -----
Rv0676c|M.tuberculosis_H37Rv -----
MMAR_1005|M.marinum_M -----
MUL_0757|M.ulcerans_Agy99 -----
MAV_2510|M.avium_104 -----
MLBr_02378|M.leprae_Br4923 -----