For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIPRDNPASRIAEATGLTPDHRISCPVGGNTPQYLVEVLGRRIWRGVSDAALIVGAESGASARKVASGSA LLEPKPIAAQDESLGDTRPGLSEAEVGAGLHWPHEVYPIFESAIAARSGRTFDEQRRWLGDLMAPFTVEA SRHLEQAWFPRARSADELSTVSPANRMVCEPYPKLLNSIIAVDMAAAFVIMAAEVAEELGVPSDRWVFPW SSATCNDVYFPVQRPDLGRSAGIRAAGKAVLAASGLHIDDIRWFDFYSCFPSAVEAAIDALDLDPADDRG FTVTGGLPYHGGPGNNYVSHSIVEMVRRCRSDPDAVGLVSGLGWYITKHSLGLWSATPPPTGWQTPDMAD AQSAIDGTSLPVASPADASGEATIDGYTVVHDRDQGPSWVPIFARMTDGRRVAARSDDAEVAVAMSRDMC VGHQVAVRQADGHVEFELS
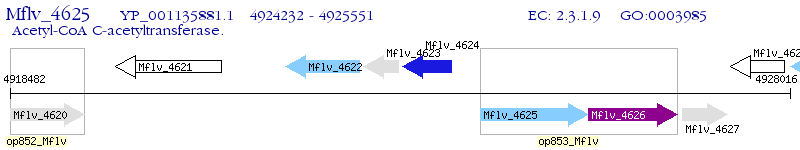
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4625 | - | - | 100% (439) | acetyl-CoA acetyltransferase |
| M. gilvum PYR-GCK | Mflv_3340 | - | 2e-67 | 37.96% (432) | acetyl-CoA acetyltransferase |
| M. gilvum PYR-GCK | Mflv_1869 | - | 3e-53 | 34.33% (434) | acetyl-CoA acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1898 | - | 4e-60 | 36.98% (411) | acetyl-CoA acetyltransferase |
| M. tuberculosis H37Rv | Rv1867 | - | 4e-60 | 36.98% (411) | acetyl-CoA acetyltransferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2042c | - | 1e-179 | 69.63% (438) | acetyl-CoA acetyltransferase |
| M. marinum M | MMAR_4547 | - | 3e-60 | 35.65% (432) | thiolase |
| M. avium 104 | MAV_2851 | - | 2e-63 | 37.10% (434) | acetyl-CoA acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_3608 | - | 3e-62 | 36.47% (414) | acetyl-CoA acetyltransferase |
| M. thermoresistible (build 8) | TH_1434 | - | 2e-60 | 41.11% (343) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4717 | - | 2e-60 | 35.65% (432) | acetyl-CoA acetyltransferase |
| M. vanbaalenii PYR-1 | Mvan_3068 | - | 3e-64 | 36.60% (418) | acetyl-CoA acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4625|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2042c|M.abscessus_ATCC_199 MPMLVEARTPVVVGVGEVTHRG-----NDFVDPIDLAVEAARRAVKDAGR
MSMEG_3608|M.smegmatis_MC2_155 ----MN-RTPVLVGYGQVSQYDENPA----TEPVDLMAEAARNAADPR--
Mvan_3068|M.vanbaalenii_PYR-1 --MPIDPRTPVLVGYGQVNQHDENPD----VEPVDLMVQAARAAADPR--
TH_1434|M.thermoresistible__bu --------------------------------------------------
Mb1898|M.bovis_AF2122/97 --MPVDPRTPVLIGYGQVNHRGDIDAEKQSIEPVDLMAAAARKAADST--
Rv1867|M.tuberculosis_H37Rv --MPVDPRTPVLIGYGQVNHRGDIDAEKQSIEPVDLMAAAARKAADST--
MAV_2851|M.avium_104 --MTVDPRTPVLIGYGQVNHRDDIDPAVRSVEPVDLMAAAVERAADAA--
MMAR_4547|M.marinum_M --MDLDPGTPVIVGVGQAVERIDDPD-YQGMSAVELAAAAAQAALEDCAA
MUL_4717|M.ulcerans_Agy99 --MDLDPGTPVIVGVGQAVERMDDPD-YQGMSAVELAAAAAQAALEDCAA
Mflv_4625|M.gilvum_PYR-GCK ---------------MIP----------RDNPASRIAEATGLTPDHRISC
MAB_2042c|M.abscessus_ATCC_199 PVERRIDTVATPGILVIP----------RDNPASRIAEAMHISPARRISC
MSMEG_3608|M.smegmatis_MC2_155 -VLQAVDSVRVVNLLSWR----------YRDPGLLLAGRIGAAGARTRYT
Mvan_3068|M.vanbaalenii_PYR-1 -LLEAVDAVRVVNLLSWR----------YRDPGLLLAQRIRADKAVTRYT
TH_1434|M.thermoresistible__bu --------------------------------------------------
Mb1898|M.bovis_AF2122/97 -VLEAVDSIRVVHMLSAH----------YRNPGQLLGERIKARTFTTGYS
Rv1867|M.tuberculosis_H37Rv -VLEAVDSIRVVHMLSAH----------YRNPGQLLGERIKARTFTTGYS
MAV_2851|M.avium_104 -VIEAVDSIRVVNILSAQ----------YRDPGLLLGQRIGARDFGTLYT
MMAR_4547|M.marinum_M DVAAAIDTVAGVRQFEISGPVPKAPLGRSNNYPRSVAQRIGADPARAILE
MUL_4717|M.ulcerans_Agy99 DVAAAIDTVAGVRQFEISGPVPKAPLGRSNNYPRSVAQRIGADPARAILE
Mflv_4625|M.gilvum_PYR-GCK PVGGNTPQYLVEVLGRRIWRGVSDAALIVGAESGASARKVASGSALL---
MAB_2042c|M.abscessus_ATCC_199 PVGGNTPQYLVEVLGGEIREGRADVVLVVGAESGHSARKLQGRALLD---
MSMEG_3608|M.smegmatis_MC2_155 GIGGNVPQTLVNHACLDLQAGRAEVVLIAGAETWRTRSRLRARGVRPDWT
Mvan_3068|M.vanbaalenii_PYR-1 GIGGNVPQTLVNMACLDIQQGKADVVLIAGAETWRTRSRIRAAGRRPDWT
TH_1434|M.thermoresistible__bu --------------------------------------------------
Mb1898|M.bovis_AF2122/97 GVGGNMPQSLVNRACLDIQRGRAGVVLLAGAETWRTRTGLRAKGSKLEWT
Rv1867|M.tuberculosis_H37Rv GVGGNMPQSLVNRACLDIQRGRAGVVLLAGAETWRTRTGLRAKGSKLEWT
MAV_2851|M.avium_104 PVGGNVPQSLVNQACLDIQRGRANVVLLAGAETWRTRRGLRAKGGKLVWT
MMAR_4547|M.marinum_M IVGGQGPQHLITELAGAIAAGRSEVAMVFGSDATSTSRHFAGRADKPDFT
MUL_4717|M.ulcerans_Agy99 IVGGQGPQHLITELAGAIAAGSSEVAMVFGSDATSTSRHFAGRADKPDFT
Mflv_4625|M.gilvum_PYR-GCK EPKPIAAQDESLGDTRPGLSEAEVGAGLHWPHEVYPIFESAIAARSGRTF
MAB_2042c|M.abscessus_ATCC_199 SPPPPRSGDESLGDARPGLSQAELSVGLSWPHEVYPIFESAIAARHGRDF
MSMEG_3608|M.smegmatis_MC2_155 RQDDTVPVAPGGDDNVPMAGDAENRIKLDRPAFVYPMFEQALRIAAGETP
Mvan_3068|M.vanbaalenii_PYR-1 KQDESVPSAPGGDESVPMAAPSDIRINLDRPAYVYPMFEQALRIAAGETP
TH_1434|M.thermoresistible__bu -----------------MATPADIRIQLDRPSYVYPLFEQALRITAGEHP
Mb1898|M.bovis_AF2122/97 VQDESVPLPDMAGDDVPMAGAAELRINLDRPAYVYPIFEQALRIAYGESI
Rv1867|M.tuberculosis_H37Rv VQDESVPLPDMAGDDVPMAGAAELRINLDRPAYVYPIFEQALRIAYGESI
MAV_2851|M.avium_104 EQDRSVPMAEVSGEDVAMAGEAEIRISLDRPAYVYPLFEQALRVSTGESV
MMAR_4547|M.marinum_M EIVDGELQDRGPGIDG-LITRYTIIHGLVDAPTQYGLLENAHRAATGLGP
MUL_4717|M.ulcerans_Agy99 EIVDGELQDRGPGIDG-LITRYTIIHGLVDAPIQYGLLENARRAATGLGP
* . * ::*.* *
Mflv_4625|M.gilvum_PYR-GCK DEQRRWLGDLMAPFTVEASRHLEQAWFPRARSADELSTVSPANRMVCEPY
MAB_2042c|M.abscessus_ATCC_199 DAQREWLGTLMAPFTAEAARHPEQAWFPRARSATELSEVTAENRMVCLPY
MSMEG_3608|M.smegmatis_MC2_155 DEHRDRIGALWSRFSAVAARN-PHAWNRDAVTAQQITQVGPDNRMISWPY
Mvan_3068|M.vanbaalenii_PYR-1 DEHRRRIGELWAQFSAVAADN-PHAWNREALTPEQIWQPSASNRMISWPY
TH_1434|M.thermoresistible__bu DAHRRRIGELWARFSAVAADN-PHAWNRKALSAEQIWRPGPDNRMISWPY
Mb1898|M.bovis_AF2122/97 ENHRKRIGELWARFSAVAADN-PHAWIRNPVTADEIWQPGPQNRMVSWPY
Rv1867|M.tuberculosis_H37Rv ENHRKRIGELWARFSAVAADN-PHAWIRNPVTADEIWQPGPQNRMVSWPY
MAV_2851|M.avium_104 EDHLTRIGRLWARFNAVAVDN-PHAWIRTPHSADDICRAGPHNRMISWPY
MMAR_4547|M.marinum_M ADYLRRMGELFAPFTKIAAGN-PFAAAPIERTIEELITVTDRNRMIAEPY
MUL_4717|M.ulcerans_Agy99 ADYLRRMGELFAPFTKIAAGN-PFAAAPIERTVEELITVTDRNRMIAEPY
:* * : *. * : * : :: ***:. **
Mflv_4625|M.gilvum_PYR-GCK PKLLNSIIAVDMAAAFVIMAAEVAEELGVPSDRWVFPWSSATCNDVYFPV
MAB_2042c|M.abscessus_ATCC_199 PKLLNSIMSVDMAAAFILMAAEVADELGVARDRWVFPWSAATCNDVYFPV
MSMEG_3608|M.smegmatis_MC2_155 PKLMNSNNMVDQGAVLILTTVQMATYLQIPMDRWVFPYAGTDSHDTYAIG
Mvan_3068|M.vanbaalenii_PYR-1 TKLMNSNNMVDQGAVLIVASAETARHLQIPSDRWVFPYAGADSHDTYAIA
TH_1434|M.thermoresistible__bu PKLMNSNNMVDQGAALILTSVQRARELRIPSERWVFPYAGTDAHDTYAIG
Mb1898|M.bovis_AF2122/97 TKLMNSNNMVDQGAALLLTSVERATRLRIPAERWVYPQAGTDAHDTPAVA
Rv1867|M.tuberculosis_H37Rv TKLMNSNNMVDQGAALLLTSVERATRLRIPAERWVYPQAGTDAHDTPAVA
MAV_2851|M.avium_104 TKLMNSNNMVDQGAALVLTSVEQARRLKVPPERWVYPLAGTDAHDTASIA
MMAR_4547|M.marinum_M PRLLVARDQVNQGAAALVMSVAAARRLGVPEEKWVYLRGHADLQEQALLQ
MUL_4717|M.ulcerans_Agy99 PRLLVARDQVNQGAAALVMSVAAARRLGVPEEKWVYLRGHADLQEQALLQ
.:*: : *: .*. :: :. * * :. ::**: . : ::
Mflv_4625|M.gilvum_PYR-GCK QRPDLGRSAGIRAAGKAVLAASGLHIDDIRWFDFYSCFPSAVEAAIDALD
MAB_2042c|M.abscessus_ATCC_199 ERPDLSRSSGIEVAARKALNAGRLTIDDIRWFDLYSCFPSAIEMTAEALD
MSMEG_3608|M.smegmatis_MC2_155 ERDELHRSPAIRIAGRRVLELAGADIDAIDFVDVYSCFPSAVQVAAAELG
Mvan_3068|M.vanbaalenii_PYR-1 ERAEFHTSPAIRIAGRRALELAGAGVDDMDLVDVYSCFPSAVQVAARELG
TH_1434|M.thermoresistible__bu ERYRLDRSPAIRIAGRRVLELAGLGIDDFRFVDVYSCFPSAVQVAAIELG
Mb1898|M.bovis_AF2122/97 DRHRLHRSTAIRIAGARALELAGLGLDDIEYVDLYSCFPSAVQVAAIELG
Rv1867|M.tuberculosis_H37Rv DRHRLHRSTAIRIAGARALELAGLGLDDIEYVDLYSCFPSAVQVAAIELG
MAV_2851|M.avium_104 ERHELHRSAAIRIAGARALQLAGLGIDDVDHVDLYSCFPSAVQVAAAELG
MMAR_4547|M.marinum_M -RPDLGHAPSAVLAAREALRVAGVGVDDIATFDLYSCFPVPVFNICDGLG
MUL_4717|M.ulcerans_Agy99 -RPDLGHAPSAVLAAREALRVAGVGVDDIATFDLYSCFPVPVFNICDGLG
* : :.. *. .* . :* . .*.***** .: *.
Mflv_4625|M.gilvum_PYR-GCK LDPAD-DRGFTVTGGLPYHGGPGNNYVSHSIVEMVRRCRSDPDAVGLVSG
MAB_2042c|M.abscessus_ATCC_199 LDPLD-DRGLTVTGGLPYHGGPGNNYVSHSIVEMVRRCRRDPTDVGLVTG
MSMEG_3608|M.smegmatis_MC2_155 LPTDDPARPLTVTGGLTFAGGPWNNYVTHSIATMAEKLVAEPGTRGLITA
Mvan_3068|M.vanbaalenii_PYR-1 LPVGKPDRPLTVTGGLTFAGGPWNNYVTHSIATMAERLAAQPGQRGLITA
TH_1434|M.thermoresistible__bu LPLDDPARPLTVTGGLTFAGGPWNNYVTHAIATMAERLIAAPGSRGLVTA
Mb1898|M.bovis_AF2122/97 LDTDDPARPLTVTGGLTFAGGPWSNYVTHSIATMAELLAANPGRRGLITA
Rv1867|M.tuberculosis_H37Rv LDTDDPARPLTVTGGLTFAGGPWSNYVTHSIATMAELLAANPGRRGLITA
MAV_2851|M.avium_104 LGLDDPARPLTVTGGLTFAGGPWSNYVMHSIATMAELLVANPGRRGLITA
MMAR_4547|M.marinum_M IATDD-PRGLTLTGGLPFFGGAGNNYSMHAVAETVARMRSEPGRFGLVGA
MUL_4717|M.ulcerans_Agy99 IATDD-PRGLTLTGGLPFFGGAGNNYSMHAVAETVARMRSEPGRFGLVGA
: . * :*:****.: **. .** *::. . * **: .
Mflv_4625|M.gilvum_PYR-GCK LGWYITKHSLGLWSATPPPTGWQTPDMADAQSAIDGTSLPVASPADASGE
MAB_2042c|M.abscessus_ATCC_199 LGWYSTKHSVGLWSATPPPTGWRLLDTAVEQAQIDSSRLGVAGVDEATGC
MSMEG_3608|M.smegmatis_MC2_155 NGGYLTKHSFGVYGTEPPEQEFRWED---VQSVVDAEPTRKL-AVEWVGT
Mvan_3068|M.vanbaalenii_PYR-1 NGGYLTKHSFGVYGTEPPKHEFRWED---VQSEVDREPTRTA-VVEWSGV
TH_1434|M.thermoresistible__bu NGGYLTKHAFGVYSTEPPPVEFRWED---VQSEVDREPVRKV-LVEWSGT
Mb1898|M.bovis_AF2122/97 NGGYLTKHSFGVYGTEPPS-EFRWED---MQPAVDREPTGDG-LVEWEGI
Rv1867|M.tuberculosis_H37Rv NGGYLTKHSFGVYGTEPPS-EFRWED---MQPAVDREPTGDG-LVEWEGI
MAV_2851|M.avium_104 NGGYLTKHSFGVYGTEPPA-EFRWED---TQSAVDREPTTDA-AVEWEGV
MMAR_4547|M.marinum_M NGGIMSKYSVGVYSTAPAQ--WQPDRSAELQTQIDSGASQPV-TEQADGP
MUL_4717|M.ulcerans_Agy99 NGGIMSKYSVGVYSTAPAQ--WQPDRSAELQTQIDSGASRPV-IEQADGP
* :*::.*::.: *. :: *. :* : *
Mflv_4625|M.gilvum_PYR-GCK ATIDGYTVVHDRDQGPSWVPIFARMTDGRRVAARSDDAEVAVAMSRDMCV
MAB_2042c|M.abscessus_ATCC_199 ATVDGYTVVYDRDGQPRWTPIIAHLSDGRRVVARSDDPQIAEAMGGEMYV
MSMEG_3608|M.smegmatis_MC2_155 GTVESWTTPFNRDGAPEKAFLAVRTPDGARTLAVITDESQAAATVREDIA
Mvan_3068|M.vanbaalenii_PYR-1 GTVESWTTPVDRDGAPEKAFLAVRTPEDARVLAVITDRAGAEATVRDDIA
TH_1434|M.thermoresistible__bu GTVESWTTPFDRDGNPEKAFVAVRTPEDSRALAVIIDPSAAAVTVSEDVA
Mb1898|M.bovis_AF2122/97 GTVEAWTTPVNRDGQPEKAFLAVRTPDGSRSLAVITDPASVQATVREDIA
Rv1867|M.tuberculosis_H37Rv GTVEAWTTPVNRDGQPEKAFLAVRTPDGSRSLAVITDPASVQATVREDIA
MAV_2851|M.avium_104 GTVEAWTTPHDREGEPEKAFLAVRTPDGSRALAVVTDRAAAQATVREDIG
MMAR_4547|M.marinum_M ATIETYTVRHD-GGRLTGIVIGRLADDGRRFLASTEDDELIGLLTDGDPL
MUL_4717|M.ulcerans_Agy99 ATIETYTVRHD-GGRLTGIVIGRLAADGRRFLASTEDDELIGLLTDGDPL
.*:: :*. : : :. * * *
Mflv_4625|M.gilvum_PYR-GCK GHQVAVRQADGHVEFELS
MAB_2042c|M.abscessus_ATCC_199 GKTVCLRNTGSFTGFELP
MSMEG_3608|M.smegmatis_MC2_155 GARVEVH-ADGTATLL--
Mvan_3068|M.vanbaalenii_PYR-1 GAAVRVH-ADGTASLE--
TH_1434|M.thermoresistible__bu GAPIRVH-ADGTATLG--
Mb1898|M.bovis_AF2122/97 GVKVAVA-PDGTATLR--
Rv1867|M.tuberculosis_H37Rv GVKVAVA-PDGTATLR--
MAV_2851|M.avium_104 GAKVAVA-ADGSAALQ--
MMAR_4547|M.marinum_M GQAIRVRSFDYGNRCVL-
MUL_4717|M.ulcerans_Agy99 GQAIRVRSFDYGNRCVL-
* : : .