For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIGLSTPVVVQTPAAASPWEAGAGIGDIARIAETADDLGFDYLTCSEHVAVPVGDAAVRGATYWDPLATL GFLAARTARIRLATSVLVLGYHHPLEIAKRYGTLDRVSGGRLILGVGVGSLQPEFDLLGAPWEERGARAD DAIRALRSSLSVAEPSYDGDFYTFGGMVVEPHAEQPRVPIWVGGRSRRSLRRAVGLADGWMPFGLRTMQI ADMLATVDIPDGFDIVLPAGPLDPREPSGCLTRLRALRDVGATAVTCSVAADSTDHYCAQLASLRGIVDE L*
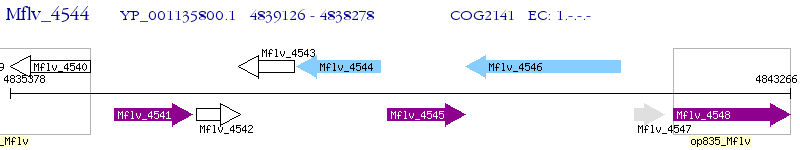
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4544 | - | - | 100% (282) | luciferase family protein |
| M. gilvum PYR-GCK | Mflv_1590 | - | 9e-38 | 36.07% (219) | luciferase family protein |
| M. gilvum PYR-GCK | Mflv_4839 | - | 1e-27 | 35.82% (201) | luciferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0978c | - | 4e-20 | 33.87% (186) | oxidoreductase |
| M. tuberculosis H37Rv | Rv0953c | - | 4e-20 | 33.87% (186) | oxidoreductase |
| M. leprae Br4923 | MLBr_00348 | - | 6e-13 | 31.00% (200) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0366 | - | 5e-30 | 36.61% (254) | hypothetical protein MAB_0366 |
| M. marinum M | MMAR_3440 | - | 9e-88 | 58.70% (276) | hypothetical protein MMAR_3440 |
| M. avium 104 | MAV_3045 | - | 8e-36 | 34.26% (216) | hypothetical protein MAV_3045 |
| M. smegmatis MC2 155 | MSMEG_2896 | - | 1e-100 | 64.03% (278) | hypothetical protein MSMEG_2896 |
| M. thermoresistible (build 8) | TH_3565 | - | 1e-106 | 65.59% (279) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_2716 | - | 5e-87 | 58.33% (276) | hypothetical protein MUL_2716 |
| M. vanbaalenii PYR-1 | Mvan_3368 | - | 1e-128 | 79.86% (278) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0978c|M.bovis_AF2122/97 -------------MHYGLVLFTSDRGITPAAAARLAESH--GFRTFYVPE
Rv0953c|M.tuberculosis_H37Rv -------------MHYGLVLFTSDRGITPAAAARLAESH--GFRTFYVPE
Mflv_4544|M.gilvum_PYR-GCK -----------MRIGLSTPVVVQTPAAASPWEAGAGIGD--IARIAETAD
Mvan_3368|M.vanbaalenii_PYR-1 ---------------MSTPVVVQTPAMASPWEGGADIED--VARIAEVAD
TH_3565|M.thermoresistible__bu -----------MRIGISTPVVMQMPGVASDWERTAGPDE--LARVAAAAD
MSMEG_2896|M.smegmatis_MC2_155 -------------MGLSTPVVVQVPQVASAWEATATVDD--LVAIAQAAD
MMAR_3440|M.marinum_M ------MSAAAAKLSIATPVVTLLPGVSAPWEKTASIED--LTQIAETAD
MUL_2716|M.ulcerans_Agy99 MLASIAMSAAAAKLSIATPVVTLLPGVSAPWEKTASIED--LTQIAETAD
MAB_0366|M.abscessus_ATCC_1997 -----------MKIGVVAPVELGVTAEPDKILEFARAAERLGFSEISVVE
MAV_3045|M.avium_104 -----------MKFAFPVPHLMRLKAIAQPWEAAVTGPE--QRRMMQCAD
MLBr_00348|M.leprae_Br4923 --------------MVERANAMKLGLQLGYWAAQPPGN---AAELVAAAE
:
Mb0978c|M.bovis_AF2122/97 HTHIPVKRQAAHPTTGDASLPDDRYMRTLDPWVSLGAASAVTSRIRLATA
Rv0953c|M.tuberculosis_H37Rv HTHIPVKRQAAHPTTGDASLPDDRYMRTLDPWVSLGAASAVTSRIRLATA
Mflv_4544|M.gilvum_PYR-GCK DLGFDYLTCSEHVAVPVGDAAV-RGATYWDPLATLGFLAARTARIRLATS
Mvan_3368|M.vanbaalenii_PYR-1 SLGFDYLTCSEHVAVPVDEAGL-RGAVYWDPLATLGFLAARTSRIRLATS
TH_3565|M.thermoresistible__bu DLGFEFLTCAEHVVVPEADAAV-RGATYWDPVATLGFLAAHTTRIRLATS
MSMEG_2896|M.smegmatis_MC2_155 ETGFDFLTCSEHVAIPRADAAR-RGAVYWDPLSTLGFLAAHTRRIRLATA
MMAR_3440|M.marinum_M KLGYYHLTCSEHIALPAAEKRR-RGTRYWDPLATLGYLAARTQRIRLATN
MUL_2716|M.ulcerans_Agy99 KLGYYHLTCSEHIALPAAEKRR-RGTRYWDPLATLSYLAARTQRIRLATN
MAB_0366|M.abscessus_ATCC_1997 HAVVIGDTQSTYPYSPTGQSHLPDDIDIPDPLELLSFVAAATTTLGLSTG
MAV_3045|M.avium_104 RWGYDMIAVPEHLVIPVDQVEL-SGSHYLHSTTAQAFIAGATERVAINSC
MLBr_00348|M.leprae_Br4923 GAGFDAVFTGEAWGS--------------DAYTPLAWWGSSTQRVRLGTS
.. . .. * : : :
Mb0978c|M.bovis_AF2122/97 VALPVEHDPITLAKSIATLDHLSHGRVSVGVGFGWNTDELVDHGVPPGRR
Rv0953c|M.tuberculosis_H37Rv VALPVEHDPITLAKSIATLDHLSHGRVSVGVGFGWNTDELVDHGVPPGRR
Mflv_4544|M.gilvum_PYR-GCK VLVLGYHHPLEIAKRYGTLDRVSGGRLILGVGVGSLQPEFDLLGAPWEER
Mvan_3368|M.vanbaalenii_PYR-1 VLVLGYHHPLEIAKRYGTLDRVSGGRLVLGVGVGSLRAEFEMLSAPWEDR
TH_3565|M.thermoresistible__bu VIVLGYHHPLEVAKRYGTVDRLSGGRVILGVGVGSLKAEFDLLDAAWSDR
MSMEG_2896|M.smegmatis_MC2_155 VLVLSYHHPLELVKRYGTLDRLSCGRLVLGVGIGSLREEFDLIGAPWEKR
MMAR_3440|M.marinum_M VLVLGYHHPLEIAKRYGTLDTVSNGRLILGVGVGSLKEEFDLIGAPFEDR
MUL_2716|M.ulcerans_Agy99 VLVLGYHHPLEIAKRYGTLDTVSNGRLILGVGVGSLKEEFDLIGAPFENR
MAB_0366|M.abscessus_ATCC_1997 VLVLPDHHPVVLAKRLATLDRLSRGRLRICAGVGWMREEIEACGGDFDRR
MAV_3045|M.avium_104 VTILPLQHPIVLAKALATADWMSGGRMMVTFGVGWLAREFELLGVPFGER
MLBr_00348|M.leprae_Br4923 VVQLSARTPTACAMAALTLDHLSGGRHILGLGVSGPQVVEGWYGQPFPKP
* : * . * * :* ** : *.. .
Mb0978c|M.bovis_AF2122/97 RTMLREYLEAMRALWTQEEAC-YDGEFVKFGPSWAWPKPVQPH-------
Rv0953c|M.tuberculosis_H37Rv RTMLREYLEAMRALWTQEEAC-YDGEFVKFGPSWAWPKPVQPH-------
Mflv_4544|M.gilvum_PYR-GCK GARADDAIRALRSSLSVAEPS-YDGDFYTFGGMVVEPHAEQPR-------
Mvan_3368|M.vanbaalenii_PYR-1 GVRADDAIRALRSSLSVSAPS-YHGEFHRYQAMMVEPHAVQPR-------
TH_3565|M.thermoresistible__bu DARADEAIRALRASLSQSRPH-HDGEHYRYSGFIVEPCAVQDR-------
MSMEG_2896|M.smegmatis_MC2_155 GARADDAIRALRASMGRAEPA-YEGPFYAFSEMTVLPHAERID-------
MMAR_3440|M.marinum_M GARADDALRALRAALSVPEPR-YHGEFYSFRDMVVDPCAVQKR-------
MUL_2716|M.ulcerans_Agy99 GARADDALRALRAALSVPEPR-YHGEFYSFRDMVVDPCAVQKR-------
MAB_0366|M.abscessus_ATCC_1997 GRAADEAIAVMRALWSTDGPTSHDGEFYSFRNAYSYPKPVRPQG------
MAV_3045|M.avium_104 GRIADEYLAAIDELWTSDTPR-FDGRYVSFADIAFEPKPVQKPH------
MLBr_00348|M.leprae_Br4923 LSRTREYIDIVRQVWAREAPVVSDGQHYPLPLTGAGATGLGKALKPITHP
: : : . .* . .
Mb0978c|M.bovis_AF2122/97 ----IPVLVGAAGTEKNFKWIARSADGWITT---PRDVDIDEPVKLLQDI
Rv0953c|M.tuberculosis_H37Rv ----IPVLVGAAGTEKNFKWIARSADGWITT---PRDVDIDEPVKLLQDI
Mflv_4544|M.gilvum_PYR-GCK ----VPIWVGGR-SRRSLRRAVGLADGWMPFGLRTMQIADMLATVDIPDG
Mvan_3368|M.vanbaalenii_PYR-1 ----VPIWVGGR-TARSLRRAVGLADGWMPFGLTPARIAEMLAAVDIPDD
TH_3565|M.thermoresistible__bu ----VPIWVGGR-TRRSLRRAVTLGDGWMPFGISAAQISEWLGSVELPDG
MSMEG_2896|M.smegmatis_MC2_155 ----VPIWVGGR-TEASLRRAVSLGDGWMPFGLRTSGIRALLDATPTPDG
MMAR_3440|M.marinum_M ----LPIWVGGR-TLRSLRRATSLAEGWVPFGISLGEARDWLSRFDIGSG
MUL_2716|M.ulcerans_Agy99 ----LPIWVGGR-TLRSLRRATSLAEGWVPFGISLGEARDWLSRFDIGSG
MAB_0366|M.abscessus_ATCC_1997 ----VPLYIGGH-SKAAARRAGRLGSGFQPLGLVGDDLTAAVGVMRAAAV
MAV_3045|M.avium_104 ----LPVWIGGD-ADAALRRAARFGSGWWPFLTPPEQIAEKLDYIKSQPC
MLBr_00348|M.leprae_Br4923 LRADIPIMLGAE-GPKNVALAAEICDGWLPIFFSPRMADMYNEWLDEGFA
:*: :*. .*: .
Mb0978c|M.bovis_AF2122/97 WAA------------------AGRDGLPQIVALDVKPVPDKLARWA----
Rv0953c|M.tuberculosis_H37Rv WAA------------------AGRDGLPQIVALDVKPVPDKLARWA----
Mflv_4544|M.gilvum_PYR-GCK FDI------------------VLPAG-PLDPR-EPSGCLTRLRALR----
Mvan_3368|M.vanbaalenii_PYR-1 FDI------------------VLAVG-PLDPR-ARDECLNRLRALR----
TH_3565|M.thermoresistible__bu FEV------------------VLATS-ALDPLGDPDAARRRLDRLR----
MSMEG_2896|M.smegmatis_MC2_155 FEV------------------VLSTGHAVDPIGDPAGTRKRLSGLA----
MMAR_3440|M.marinum_M FEV------------------VLSSPEPLNPIGDPERTRHLLADLA----
MUL_2716|M.ulcerans_Agy99 FEV------------------VLSSPEPLNPIGDPERTRHLLADLA----
MAB_0366|M.abscessus_ATCC_1997 DAG------------------RDPRGLDLVLGHGLHRVDQAALDQA----
MAV_3045|M.avium_104 YDGRPFEVMHGLGTNRVGEGHVARRDVRDRPGMSAGQIVDRLGWLA----
MLBr_00348|M.leprae_Br4923 RTG----------------ARRSREDFEICASAQIVVTEDRAAAFAGIKP
Mb0978c|M.bovis_AF2122/97 -------ELGVTEVLFGMPDRSADDAAAYVERLAAKLACCV---------
Rv0953c|M.tuberculosis_H37Rv -------ELGVTEVLFGMPDRSADDAAAYVERLAAKLACCV---------
Mflv_4544|M.gilvum_PYR-GCK -------DVGATAVTCSVAADSTDHYCAQLASLRGIV--DEL--------
Mvan_3368|M.vanbaalenii_PYR-1 -------DVGATAVTCSLSASSAEHYCDQLASLRDLV--DEL--------
TH_3565|M.thermoresistible__bu -------ELGATAVTATVSATSAAHYCEQLAALRDLAMGDAL--------
MSMEG_2896|M.smegmatis_MC2_155 -------DAGATVASCTISASSAAHYRDQVARLSEIAGTVEGRGA-----
MMAR_3440|M.marinum_M -------AHGATTICATFAHSSRQQYLDNLHALAELDSP-----------
MUL_2716|M.ulcerans_Agy99 -------AHGATTICATFAHSSRQQYLDNLHALAELDSP-----------
MAB_0366|M.abscessus_ATCC_1997 -------AHAGAGRMLLSASRRATTLEAVLEEMAGCATRLELGGPR----
MAV_3045|M.avium_104 -------EQGVTVTAVPVPAVGGVDEYLDYAQWVIEEIKPQLA-------
MLBr_00348|M.leprae_Br4923 FLALYMGGMGAEGTNFHADVYRRMGYAEVVDEVTALFRSNQKDKAAEIIP
.
Mb0978c|M.bovis_AF2122/97 ------------------------------------------------
Rv0953c|M.tuberculosis_H37Rv ------------------------------------------------
Mflv_4544|M.gilvum_PYR-GCK ------------------------------------------------
Mvan_3368|M.vanbaalenii_PYR-1 ------------------------------------------------
TH_3565|M.thermoresistible__bu ------------------------------------------------
MSMEG_2896|M.smegmatis_MC2_155 ------------------------------------------------
MMAR_3440|M.marinum_M ------------------------------------------------
MUL_2716|M.ulcerans_Agy99 ------------------------------------------------
MAB_0366|M.abscessus_ATCC_1997 ------------------------------------------------
MAV_3045|M.avium_104 ------------------------------------------------
MLBr_00348|M.leprae_Br4923 NELVDSTMIVGDVDYVCKQIAAWSAAGVTMMMVSASSVEQVRDLAGLF