For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSDPQTLRALAGLPLTPMSLADSALVLIDCQNTYTRGVMELDGVQAALDEAATLLDRARTAGIAVVHIQH DDGPGSLYDIEGESGAIVDRVAPRDGERVVVKNYPNSFVQTDLDDVLKSYGASNLVLAGFMTHMCVNSTA RGAFNLGYAPTVVAAATATRALPGVDGDTVAASAVHAASLAAVADLFAVVVPGEKDIPG
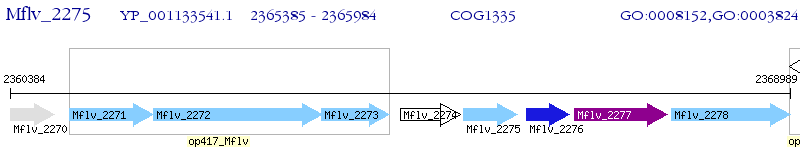
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2275 | - | - | 100% (199) | isochorismatase hydrolase |
| M. gilvum PYR-GCK | Mflv_4511 | - | 5e-06 | 27.27% (132) | isochorismatase hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3431c | - | 7e-10 | 28.82% (170) | isochorismatase hydrolase |
| M. marinum M | MMAR_2657 | - | 2e-08 | 27.78% (144) | isochorismatase family protein |
| M. avium 104 | MAV_1602 | - | 4e-06 | 24.56% (171) | isochorismatase family protein |
| M. smegmatis MC2 155 | MSMEG_4976 | - | 1e-77 | 75.13% (193) | isochorismatase hydrolase |
| M. thermoresistible (build 8) | TH_2198 | - | 8e-08 | 28.15% (135) | isochorismatase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4420 | - | 3e-90 | 85.35% (198) | isochorismatase hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2275|M.gilvum_PYR-GCK -----------------------------------MSDPQTLRALAGLPL
Mvan_4420|M.vanbaalenii_PYR-1 -----------------------------------MSDLPTLRSLSGLPL
MSMEG_4976|M.smegmatis_MC2_155 -----------------------------------MTR-PTLREISSLPA
MAB_3431c|M.abscessus_ATCC_199 MTGQVDAQPYPWPFDGPVDPGRTAVLCIDWQVDFCGRGGYVDTMGYDLSL
MMAR_2657|M.marinum_M -------------------------------------------MGYDLSL
MAV_1602|M.avium_104 ----------MTDLAELVAPAHTAVITQEVQG--AVVGPDAGLGALAAEA
TH_2198|M.thermoresistible__bu ---------------------------------------MSDTAVVVVDM
Mflv_2275|M.gilvum_PYR-GCK TPMSLADSALVLIDCQNTYTRGVMELDGVQAALDEAATLLDRARTAGIAV
Mvan_4420|M.vanbaalenii_PYR-1 QPVSLADSALVLIDCQNTYTYGVMELEGVQAALDEAAALLDRARTAGIPV
MSMEG_4976|M.smegmatis_MC2_155 EPVRLADSALVLIDCQNTYTRGVMELEGVQAALDATAELLDRARTAGIPI
MAB_3431c|M.abscessus_ATCC_199 TRAGLEPTARVLAAAR---AVGMTIIHTREGHRPDLSDLPANKRWRSARI
MMAR_2657|M.marinum_M VRAPLGPAQTVLEAAR---KLGLLIVHTREGHRPDLSDLPANKRWRSARI
MAV_1602|M.avium_104 RRVALPNIVRLLPPAR---AAGVRIVHCLVQRRPD-----GLGSNHNAKI
TH_2198|M.thermoresistible__bu FNTYDHEDAELLTPNVESILDPLAKLIAEARHRDDVDLIYVNDNHG----
:* : :
Mflv_2275|M.gilvum_PYR-GCK VHIQHDDGPGSLYDIEGESG-AIVDRVAPRDGERVVVKNY-PNSFVQTDL
Mvan_4420|M.vanbaalenii_PYR-1 VHIQHDDGPGSPYDITAEIG-AIVDRVAPRGDEPVVVKNY-PNSFVQTDL
MSMEG_4976|M.smegmatis_MC2_155 IHIQHDSGPGSPYDVREEIG-AIVESVAPRGDEPVVVKNF-PNSFVQTDL
MAB_3431c|M.abscessus_ATCC_199 GAEIGVAGPCGRILVKGEPGWEIVPEVAPLPGEPIIDKPG-KGAFYATDL
MMAR_2657|M.marinum_M GAEIGVAGPCGRVLTRGEPGWEIIPEMEPLPGEMVVDKLG-KGSFYATDL
MAV_1602|M.avium_104 FALGRRSGQGRVDISPGTPGATLLPELGPAPSDLVLSRWHGVGPMGGTDL
TH_2198|M.thermoresistible__bu -DFTAEFSDVVRTALDGARP-DLVRPIVPDDGCRALLKVR-HSVFYATAL
. :: : * . : : . : * *
Mflv_2275|M.gilvum_PYR-GCK DDVLKSYGASNLVLAGFMTHMCVNSTARGAFNLGYAPTVVAAATATRALP
Mvan_4420|M.vanbaalenii_PYR-1 DDRLKAVDASNLVLAGFMTHMCVNSTARGAFNLGYAPTVVAAATATRALP
MSMEG_4976|M.smegmatis_MC2_155 QQRLQAVNASNLVLAGFMTHMCVNSTARGAFNLGFAPTVVAAATATRSLP
MAB_3431c|M.abscessus_ATCC_199 DLVLRTRGIRYIVLTGITTDVCVHTTMREANDRGFECLILSDCTGATDAG
MMAR_2657|M.marinum_M ELILTTRRITHLIFTGIATDVCVHTTMREANDRGYECLLLSDCTGATDYA
MAV_1602|M.avium_104 DAVLRNLGVSTLVVVGVSLNIAIPNVVMDAVNAAYRVVVPRDAVAG--VP
TH_2198|M.thermoresistible__bu EYLLTRLSARRVILTGQVTEQCILYSALDAYVRHFEVVVPPDAVAHIDAG
: * ::..* . .: * : : ...
Mflv_2275|M.gilvum_PYR-GCK GVDGDTVAASAVHAASLAAVADLFAVVVPGEKDIPG--
Mvan_4420|M.vanbaalenii_PYR-1 GVGGEDVPAAAVHSASLAAVADLFAVVVPGARDIPD--
MSMEG_4976|M.smegmatis_MC2_155 GPDGNLVPASAMQSASLAAIADLFAVVVPDVGSIPD--
MAB_3431c|M.abscessus_ATCC_199 N-HAAALKMVTMQGGVFGAVADSNQLLGAL--------
MMAR_2657|M.marinum_M N-HLAALKMITMQGGVFGAHATNGALLEAFNKVRDRRG
MAV_1602|M.avium_104 AEYGEAVIANTLS--LLATITTTDELLRAWSRP-----
TH_2198|M.thermoresistible__bu LGEAALQMMARNMSAELPPSADCLP-------------
: . :