For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSTQSVSPAVLFDIDGTLVDSNYLHVHAWMRAFDDENLPVSAWRIHRCIGMDGTTLVRTLSGDAGQGVLD RLKDAHSRYYKENSSLLAPLPGARDLLRRVAATGLQVVLATSAPDDELTLLREVLDCDDVVAAVTSSADV DTAKPKPDIVQVALDRAGVGAQDAVFVGDAVWDCEAAGRAGVTSIGLLSGGVSRGELREAGAAGVFEDAG DLLAKLDSTVIGDLAARVSDPR
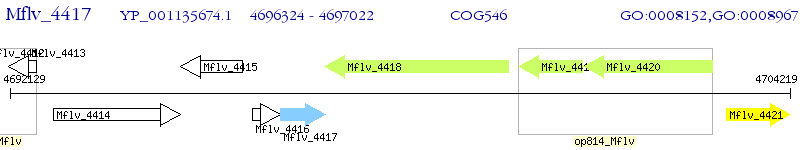
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4417 | - | - | 100% (232) | HAD family hydrolase |
| M. gilvum PYR-GCK | Mflv_2889 | - | 3e-08 | 25.23% (218) | HAD family hydrolase |
| M. gilvum PYR-GCK | Mflv_0023 | - | 3e-05 | 32.93% (82) | HAD family hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3410 | - | 6e-05 | 29.51% (122) | hypothetical protein Mb3410 |
| M. tuberculosis H37Rv | Rv3376 | - | 6e-05 | 29.51% (122) | hypothetical protein Rv3376 |
| M. leprae Br4923 | MLBr_00393 | - | 2e-06 | 25.42% (240) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_1901c | - | 4e-07 | 26.92% (182) | hypothetical protein MAB_1901c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0437 | - | 1e-78 | 70.05% (207) | hydrolase |
| M. smegmatis MC2 155 | MSMEG_4401 | - | 8e-09 | 32.81% (128) | phosphonoacetaldehyde hydrolase |
| M. thermoresistible (build 8) | TH_3516 | - | 4e-10 | 29.41% (153) | PUTATIVE HAD-superfamily hydrolase, subfamily IA, variant 3 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1937 | - | 1e-101 | 78.60% (229) | HAD family hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4417|M.gilvum_PYR-GCK ---------------MSTQSVSPAVLFDIDGTLVDSNYLHVHAWMRAFDD
Mvan_1937|M.vanbaalenii_PYR-1 -----------MPRPMSIEKPSPAVLFDVDGTLVDSNYLHVYAWLRAFDA
MAV_0437|M.avium_104 ---------------------------------MDSNYLHVHSWQRAFDD
MSMEG_4401|M.smegmatis_MC2_155 ------MNISEGTRMSSTVRRPELVVLDIAGTTVDEGSAVYRVLEQTVVA
TH_3516|M.thermoresistible__bu ----------------------------MAGTTVTDDGLVLRAFDAAATA
Mb3410|M.bovis_AF2122/97 -------------------MSISAVVFDRDGVLTSFDWTRAEEDVRRITG
Rv3376|M.tuberculosis_H37Rv -------------------MSISAVVFDRDGVLTSFDWTRAEEDVRRITG
MLBr_00393|M.leprae_Br4923 MANWDHPTSKVRSTVLGLPEQVRACLFDLDGVLTDTASVHAKAWQTMFDT
MAB_1901c|M.abscessus_ATCC_199 ----------------------MLVIFDLDGTLTDSAPGIVSSFRHALSH
Mflv_4417|M.gilvum_PYR-GCK ENLPVSAWR-------------IHRCIGMDGTTLVRTLSG----DAGQGV
Mvan_1937|M.vanbaalenii_PYR-1 ESVPVDSWR-------------IHRCIGMDGTTLVRALAG----DASERV
MAV_0437|M.avium_104 EDIAVASWQ-------------IHRCIGMDGSTLVRTLSG----DAPDDV
MSMEG_4401|M.smegmatis_MC2_155 HGGTPSEAD-------------IAKWHGAAKHEALRALLAPADDAALQVI
TH_3516|M.thermoresistible__bu VGMAESGAERE------AARRYVVDTMGRSKIEVFRALLA--DEDRAQRA
Mb3410|M.bovis_AF2122/97 LPLEEIERRWG------GWLNGLTIDDAFVETQPISEFLS----SLAREL
Rv3376|M.tuberculosis_H37Rv LPLEEIERRWG------GWLNGLTIDDAFVETQPISEFLS----SLAREL
MLBr_00393|M.leprae_Br4923 YLYQRAKHTGENFVPFDPTADYRQYVDGKKREDGVRSFLGSRGITLPEGS
MAB_1901c|M.abscessus_ATCC_199 VGAPEPTGD-------------IASRVVGPPMHITLGSMG-----LGERA
.
Mflv_4417|M.gilvum_PYR-GCK LDRLKDAHSRYYKENSSL-----------LAPLPGARDLLRRVAATGLQV
Mvan_1937|M.vanbaalenii_PYR-1 LDTLKDRHSEFYRETASL-----------LAPLPGARDLLRRVAGLGLQV
MAV_0437|M.avium_104 GERLRDRHSRYYRELTPL-----------LKPLPGARAMLRRVADLGLQV
MSMEG_4401|M.smegmatis_MC2_155 VEDFRARLKAAYAEHP-------------PVPLPGVPDAIRDLRASGIKV
TH_3516|M.thermoresistible__bu NAAFESAYDRFIAEG--------------VAPVPGAAEAIGELRSAGVKV
Mb3410|M.bovis_AF2122/97 ELGSKARDELVRLDYMAF-----------AQGYPDARPALEEARRRGLKV
Rv3376|M.tuberculosis_H37Rv ELGSKARDELVRLDYMAF-----------AQGYPDARPALEEARRRGLKV
MLBr_00393|M.leprae_Br4923 ADNSGDVETIYGLGNRKDDLFRQVLKEHGIEVFDGSRRYLEAITYAGLGV
MAB_1901c|M.abscessus_ATCC_199 DDAIAAYRADYGERGWAVN-----------EMFDGIRDVLTRLREAGVPM
. : *: :
Mflv_4417|M.gilvum_PYR-GCK VLATSAPDDELTLLREVLDCD---DVVAAVTSSADVDTAKPKPDIVQVAL
Mvan_1937|M.vanbaalenii_PYR-1 VLATSAPEDELRMLREVLDCD---DAVAAVTSSQDVDTAKPQPDIVAVAL
MAV_0437|M.avium_104 VLASSAPEDELEILRKVLDCD---DVIAATTSSRDVDTAKPEPGIVRVAL
MSMEG_4401|M.smegmatis_MC2_155 ALNTGFDRDIVDALLASLGWDGD-SVVDTVVCGSDVAAGRPAPYMIYRAM
TH_3516|M.thermoresistible__bu ALTTGFSPATQARILDALGWT---DIADLTLAPDTGVRGRPYPDLVLTAV
Mb3410|M.bovis_AF2122/97 GVLTNN--SLLVSARSLLQCAALHDLVDVVLSSQMIGAAKPDPRAYQAIA
Rv3376|M.tuberculosis_H37Rv GVLTNN--SLLVSARSLLQCAALHDLVDVVLSSQMIGAAKPDPRAYQAIA
MLBr_00393|M.leprae_Br4923 AVVSSSTNTRDVLKITGLDRFVQ-QQVDGITLREEHIAGKPAPDSYLRGA
MAB_1901c|M.abscessus_ATCC_199 VVATSKSEPIAARILEHFGLA---EFFQVIAGASPDGVRSSKADVIKHAL
: :. : . . .
Mflv_4417|M.gilvum_PYR-GCK DRAGVGAQDAVFVG-DAVWDCEAAGRAGVT-SIGLLSGG--VSRGELREA
Mvan_1937|M.vanbaalenii_PYR-1 DRAGVTADDAVFVG-DAVWDCEAARRAGVP-SIGVLSGG--VSRAELREA
MAV_0437|M.avium_104 DRAGVRAEEAVFVG-DAVWDAHAARGAGLA-CIGLCSGG--IARDELRAA
MSMEG_4401|M.smegmatis_MC2_155 EHSGVQDISRVLVAGDTPRDLQAGINAGAGYVVGVLSGA--SDEAELSAH
TH_3516|M.thermoresistible__bu LRVGIDDVRHVAVAGDTASDIETGLRAGASIATGVLTGA--HDEQTLRAA
Mb3410|M.bovis_AF2122/97 EALGVSTTSCLFFD-DIADWVEGARCAGMRAYLVDRSGQ--TRDGVVRDL
Rv3376|M.tuberculosis_H37Rv EALGVSTTSCLFFD-DIADWVEGARCAGMRAYLVDRSGQ--TRDGVVRDL
MLBr_00393|M.leprae_Br4923 QLLDVAPDAAAVFE-DALSGVQAGLSGHFGFVVGINRTGRVDQAEELRRI
MAB_1901c|M.abscessus_ATCC_199 SQLDDVPPTGVVMVGDRVHDVEGAAAHGIGTVVVDWGYG-ATDFEGDPGQ
. . * . .
Mflv_4417|M.gilvum_PYR-GCK GAAGVFEDAGDLLAKLDSTVIGDLAARVSDPR
Mvan_1937|M.vanbaalenii_PYR-1 GARDVFEDAAELLTELDSTVVGELAGRVSG--
MAV_0437|M.avium_104 GADPVFADPQDLLDHLESTRIAALARGG----
MSMEG_4401|M.smegmatis_MC2_155 PHTHLLSSVADLPELFGVTRTAATAS------
TH_3516|M.thermoresistible__bu GATHVLESVADLPRL--VLR------------
Mb3410|M.bovis_AF2122/97 SSLGAILDGAGP--------------------
Rv3376|M.tuberculosis_H37Rv SSLGAILDGAGP--------------------
MLBr_00393|M.leprae_Br4923 GANVVVTDLAELL-------------------
MAB_1901c|M.abscessus_ATCC_199 LAEWIIGRVSTPEQLLEELDV-----------
.