For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DSNYLHVHSWQRAFDDEDIAVASWQIHRCIGMDGSTLVRTLSGDAPDDVGERLRDRHSRYYRELTPLLKP LPGARAMLRRVADLGLQVVLASSAPEDELEILRKVLDCDDVIAATTSSRDVDTAKPEPGIVRVALDRAGV RAEEAVFVGDAVWDAHAARGAGLACIGLCSGGIARDELRAAGADPVFADPQDLLDHLESTRIAALARGG*
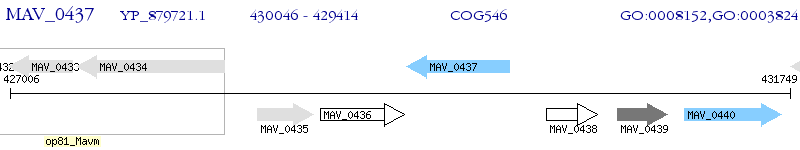
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0437 | - | - | 100% (210) | hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4417 | - | 9e-79 | 70.05% (207) | HAD family hydrolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4401 | - | 6e-08 | 30.00% (160) | phosphonoacetaldehyde hydrolase |
| M. thermoresistible (build 8) | TH_2509 | - | 1e-06 | 26.49% (151) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1937 | - | 1e-76 | 67.63% (207) | HAD family hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4417|M.gilvum_PYR-GCK ---------MSTQSVSPAVLFDIDGTLVDSNYLHVHAWMRAFDDE--NLP
Mvan_1937|M.vanbaalenii_PYR-1 -----MPRPMSIEKPSPAVLFDVDGTLVDSNYLHVYAWLRAFDAE--SVP
MAV_0437|M.avium_104 ---------------------------MDSNYLHVHSWQRAFDDE--DIA
MSMEG_4401|M.smegmatis_MC2_155 MNISEGTRMSSTVRRPELVVLDIAGTTVDEGSAVYRVLEQTVVAH--GGT
TH_2509|M.thermoresistible__bu -LTFDRESPQPDRPGIRGVVFDKDGTLFDFTRTWTPWCEQLVGELSGGDP
.* : . . .
Mflv_4417|M.gilvum_PYR-GCK VSAWRIHRCIGMDGTTLVR----TLSGDAGQGVLDRLK---------DAH
Mvan_1937|M.vanbaalenii_PYR-1 VDSWRIHRCIGMDGTTLVR----ALAGDASERVLDTLK---------DRH
MAV_0437|M.avium_104 VASWQIHRCIGMDGSTLVR----TLSGDAPDDVGERLR---------DRH
MSMEG_4401|M.smegmatis_MC2_155 PSEADIAKWHGAAKHEALR----ALLAPADDAALQVIV---------EDF
TH_2509|M.thermoresistible__bu GLARRLGAALGFDLELRMFEPDSVVLTDTIQAVVKTLLGVLPPDTDADRL
: * : .: : : . . : :
Mflv_4417|M.gilvum_PYR-GCK SRYYKEN--SSLLAPLPGARDLLRRVAATGLQVVLATSAPDDELTLLREV
Mvan_1937|M.vanbaalenii_PYR-1 SEFYRET--ASLLAPLPGARDLLRRVAGLGLQVVLATSAPEDELRMLREV
MAV_0437|M.avium_104 SRYYREL--TPLLKPLPGARAMLRRVADLGLQVVLASSAPEDELEILRKV
MSMEG_4401|M.smegmatis_MC2_155 RARLKAAYAEHPPVPLPGVPDAIRDLRASGIKVALNTGFDRDIVDALLAS
TH_2509|M.thermoresistible__bu FRHVDDSIATRSPAQVVPLRPLLERLRSAGIVLGLATNDQKK--STLRHL
: :. : *: : * :. . *
Mflv_4417|M.gilvum_PYR-GCK L--DCDDVVAAVTSSADVDTAKPKPDIVQVALDRAGVGAQDAVFVG-DAV
Mvan_1937|M.vanbaalenii_PYR-1 L--DCDDAVAAVTSSQDVDTAKPQPDIVAVALDRAGVTADDAVFVG-DAV
MAV_0437|M.avium_104 L--DCDDVIAATTSSRDVDTAKPEPGIVRVALDRAGVRAEEAVFVG-DAV
MSMEG_4401|M.smegmatis_MC2_155 LGWDGDSVVDTVVCGSDVAAGRPAPYMIYRAMEHSGVQDISRVLVAGDTP
TH_2509|M.thermoresistible__bu QQAGIDSCLSFVADADSGHGIKPGPGMLVAFCEHTGLSPQHCAMVG-DSV
. *. : .. . . :* * :: :::*: .:*. *:
Mflv_4417|M.gilvum_PYR-GCK WDCEAAGRAGVT-SIGLLSGGVSRGELREAGAAGVFEDAGDLLAKLDSTV
Mvan_1937|M.vanbaalenii_PYR-1 WDCEAARRAGVP-SIGVLSGGVSRAELREAGARDVFEDAAELLTELDSTV
MAV_0437|M.avium_104 WDAHAARGAGLA-CIGLCSGGIARDELRAAGADPVFADPQDLLDHLESTR
MSMEG_4401|M.smegmatis_MC2_155 RDLQAGINAGAGYVVGVLSGASDEAELSAHPHTHLLSSVADLPELFGVTR
TH_2509|M.thermoresistible__bu FDMLAAQSANFL-RIGVCSGPTPAERLXXX---LLRTGRPWVPVCWHREH
* *. *. :*: ** .* : . :
Mflv_4417|M.gilvum_PYR-GCK IGDLAARVSDPR
Mvan_1937|M.vanbaalenii_PYR-1 VGELAGRVSG--
MAV_0437|M.avium_104 IAALARGG----
MSMEG_4401|M.smegmatis_MC2_155 TAATAS------
TH_2509|M.thermoresistible__bu TSDRGGRRGSR-
. .