For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GDAVSGPAIRVRGLRYSYPDGHVALDGIDLDIADGERVALLGPNGAGKTTLMLHLNGVLRASAGDVEIGG TVLSRSTLRDIRRRVGLVFQDPDDQLFMPTLAQDVAFGPANFGVRGAELDARVAHALAVVSMTELAGRSP AHMSGGQRRRAALATVLACEPEVLVLDEPSANLDPVARRELAETLSGLDATMLIVTHDLPYAAQLCRRAI VMDGGVIVADGAIDEVLSDTALLAAHRLELPWGFTVGLR*
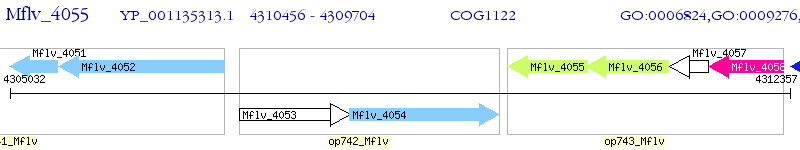
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4055 | - | - | 100% (250) | cobalt ABC transporter, ATPase subunit |
| M. gilvum PYR-GCK | Mflv_0971 | - | 6e-33 | 38.35% (206) | ABC transporter related |
| M. gilvum PYR-GCK | Mflv_3463 | - | 9e-31 | 38.43% (216) | ABC transporter related |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1270 | sugC | 5e-30 | 34.84% (221) | sugar-transport ATP-binding protein ABC transporter SugC |
| M. tuberculosis H37Rv | Rv1238 | sugC | 5e-30 | 34.84% (221) | sugar-transport ATP-binding protein ABC transporter SugC |
| M. leprae Br4923 | MLBr_01089 | - | 4e-29 | 34.11% (214) | putative ABC-transport protein, ATP-binding component |
| M. abscessus ATCC 19977 | MAB_4237c | - | 8e-28 | 34.32% (236) | amino acid ABC transporter ATP-binding protein |
| M. marinum M | MMAR_1901 | - | 4e-28 | 35.51% (214) | ABC-type sugar transport protein, ATPase component |
| M. avium 104 | MAV_1377 | sugC | 2e-29 | 34.84% (221) | ABC transporter, ATP-binding protein SugC |
| M. smegmatis MC2 155 | MSMEG_2610 | - | 1e-103 | 76.57% (239) | cobalt transport protein ATP-binding subunit |
| M. thermoresistible (build 8) | TH_2525 | - | 3e-41 | 40.43% (230) | PUTATIVE putative cobalt ABC transporter, ATPase subunit |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 1e-27 | 37.39% (230) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_2294 | - | 1e-117 | 86.48% (244) | cobalt ABC transporter, ATPase subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4237c|M.abscessus_ATCC_199 MS-------TIEVRGVRKSYG------QVTALRDVNLTVHQGEVTVIIGP
MUL_3658|M.ulcerans_Agy99 MTGPGTGGHAIVVRDAFKQYG------DFVALDHVDFVVPAGSLTALLGP
Mb1270|M.bovis_AF2122/97 -------MAEIVLDHVNKSYP-----DGHTAVRDLNLTIADGEFLILVGP
Rv1238|M.tuberculosis_H37Rv -------MAEIVLDHVNKSYP-----DGHTAVRDLNLTIADGEFLILVGP
MAV_1377|M.avium_104 -------MAEIVLDHVSKSYP-----DGATAVRDLNLTIADGEFLILVGP
MLBr_01089|M.leprae_Br4923 -------MAEIVLEHVNKNYP-----NGATAVHDLSITVADGEFLILIGP
MMAR_1901|M.marinum_M -------MAKVQYSGVTYRYPG----SGAPAVNNLDLEIADGEFLVLVGP
Mflv_4055|M.gilvum_PYR-GCK ---MGDAVSGPAIRVRGLRYS---YPDGHVALDGIDLDIADGERVALLGP
Mvan_2294|M.vanbaalenii_PYR-1 -------MTAPAIRVSGLHYA---YPGGHEALGGIDLEVAAGERVALLGP
MSMEG_2610|M.smegmatis_MC2_155 -------MTDTAISIAALCHV---YPDGHIGLDGVDLEVAEGERVAVLGP
TH_2525|M.thermoresistible__bu -------VSDDDPALLALRDAGFRYPRGPRVLDGVQLSIGPGQRLALVGA
: :.: : *. ::*.
MAB_4237c|M.abscessus_ATCC_199 SGSGKSTLLRALNHLEKVDAGTIRIDGELIGYRQRGSVLYELPERAILRQ
MUL_3658|M.ulcerans_Agy99 SGSGKSTLLRTIAGLDQPDSGNITINGRDVTR--------------VPPQ
Mb1270|M.bovis_AF2122/97 SGCGKTTTLNMIAGLEDISSGELRIAGERVNEK--------------APK
Rv1238|M.tuberculosis_H37Rv SGCGKTTTLNMIAGLEDISSGELRIAGERVNEK--------------APK
MAV_1377|M.avium_104 SGCGKTTTLNMIAGLEDISSGELRIGGERVNEK--------------APK
MLBr_01089|M.leprae_Br4923 SGCGKTTTLNMIAGLEDISSGELRIDGDRVNEK--------------APK
MMAR_1901|M.marinum_M SGCGKSTILRVLAGLESVESGRITIGDTDVTDL--------------PPR
Mflv_4055|M.gilvum_PYR-GCK NGAGKTTLMLHLNGVLRASAGDVEIGGTVLS------------RSTLRDI
Mvan_2294|M.vanbaalenii_PYR-1 NGAGKTTLMLHLNGVLTALQGSVEIGGTMLS------------RNTLRDI
MSMEG_2610|M.smegmatis_MC2_155 NGAGKTTLMLHLNGVLTATSGTVRICGVPVA------------RNTLRDI
TH_2525|M.thermoresistible__bu NGSGKTTLLRLLVGLAKPTSGTLLLDGNVVTGS----------RADRRRL
.*.**:* : : : * : : . :
MAB_4237c|M.abscessus_ATCC_199 RSQVGFVFQN-FNLFPHLTVLENVIEAPLAARRAPRAELEAEATRLLERV
MUL_3658|M.ulcerans_Agy99 RRGIGFVFQH-YAAFKHLTVRDNVAYG-LKIRKRPKAEIRDKVDNLLEVV
Mb1270|M.bovis_AF2122/97 DRDIAMVFQS-YALYPHMTVRQNIAFP-LTLAKMRKADIAQKVSETAKIL
Rv1238|M.tuberculosis_H37Rv DRDIAMVFQS-YALYPHMTVRQNIAFP-LTLAKMRKADIAQKVSETAKIL
MAV_1377|M.avium_104 DRDIAMVFQS-YALYPHMTVRQNIAFP-LTLAKMKKPEIAQKVAETAKIL
MLBr_01089|M.leprae_Br4923 DRDIAMVFQS-YALYPHMTVRQNIAFP-LMLAKVKKAEIAQKVSETAQIL
MMAR_1901|M.marinum_M ARDVAMVFQN-YALYPNMTVAQNMGFA-LRNAGMSRADTRQRVLEVASML
Mflv_4055|M.gilvum_PYR-GCK RRRVGLVFQDPDDQLFMPTLAQDVAFG-PANFGVRGAELDARVAHALAVV
Mvan_2294|M.vanbaalenii_PYR-1 RRRVGLVFQDPDDQLFMPTLAQDVAFG-PANFGIRGAELEARVARALEVV
MSMEG_2610|M.smegmatis_MC2_155 RRRVGLVFQDPDDQLFMPTVAQDVAFG-PANFGLAGEELAARVRHALELV
TH_2525|M.thermoresistible__bu RTRVQMVLQEPDDQIIGATVRADVSFG-PLNLGLDHTEVHDRVDAAMAAL
: :*:* *: :: : .. :
MAB_4237c|M.abscessus_ATCC_199 GVADKAAQYPRRLSGGQQQRVAIARALALRPKVILFDEPTSALDPELVTE
MUL_3658|M.ulcerans_Agy99 GLSGFHGRYPDQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVRED
Mb1270|M.bovis_AF2122/97 DLTNLLDRKPSQLSGGQRQRVAMGRAIVRHPKAFLMDEPLSNLDAKLRVQ
Rv1238|M.tuberculosis_H37Rv DLTNLLDRKPSQLSGGQRQRVAMGRAIVRHPKAFLMDEPLSNLDAKLRVQ
MAV_1377|M.avium_104 DLTDFLDRKPSQLSGGQRQRVAMGRAIVRHPKAFLMDEPLSNLDAKLRVQ
MLBr_01089|M.leprae_Br4923 DLTDLLDRKPSQLSGGQRQRVAMGRAIVRHPKAFLMDEPLSNLDAKLRVR
MMAR_1901|M.marinum_M ELTELLDRKPSKLSGGQRQRVAMGRAIVRRPRVFCMDEPLSNLDAKLRVS
Mflv_4055|M.gilvum_PYR-GCK SMTELAGRSPAHMSGGQRRRAALATVLACEPEVLVLDEPSANLDPVARRE
Mvan_2294|M.vanbaalenii_PYR-1 SMTELAARSPAHMSGGQRRRAALATVLACEPEVLVLDEPSANLDPVARRE
MSMEG_2610|M.smegmatis_MC2_155 SLTEHADRSPAHLSGGQRRRAALATVLACDAEILVLDEPSANLDPVARRE
TH_2525|M.thermoresistible__bu GITELGDRTPHHLSFGQRKRVAIAGAVAMRPSVLLLDEATAGLDPRAVED
:: : * ::* **::* *:. .:. . : :**. . **.
MAB_4237c|M.abscessus_ATCC_199 VLDVIAQLARDG-ATLVIVTHEIGLAREIADTVVFMDRGAIVEQGPPKEV
MUL_3658|M.ulcerans_Agy99 LRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEV
Mb1270|M.bovis_AF2122/97 MRGEIAQLQRRLGTTTVYVTHDQTEAMTLGDRVVVMYGGIAQQIGTPEEL
Rv1238|M.tuberculosis_H37Rv MRGEIAQLQRRLGTTTVYVTHDQTEAMTLGDRVVVMYGGIAQQIGTPEEL
MAV_1377|M.avium_104 MRGEIARLQKRLGTTTVYVTHDQTEAMTLGDRVVVMHSGVAQQIGTPDEL
MLBr_01089|M.leprae_Br4923 TRGEIARLQRRLGATTVYVTHDQTEAMTLGDRVVVMRSGVVQQIGTPDEL
MMAR_1901|M.marinum_M TRSHISGLQRQLGTTTVYVTHDQVEAMTMGHRVAVLKDGVLQQVDTPREL
Mflv_4055|M.gilvum_PYR-GCK LAETLSGLDAT----MLIVTHDLPYAAQLCRRAIVMDGGVIVADGAIDEV
Mvan_2294|M.vanbaalenii_PYR-1 LAETLSGLDAT----MVIVTHDLPYAAQLCQRAIVMDHGVIVADGTTADV
MSMEG_2610|M.smegmatis_MC2_155 LAETLAALDVT----MLIVTHDLPYAAQLCTRAVVIDDGRVVADGDIGEI
TH_2525|M.thermoresistible__bu LLRTLTELSDAG-TAVVLATHDVDVAWRWSQETLVLHDG-TVRRGPTHTL
: * : .**: * .: * . :
MAB_4237c|M.abscessus_ATCC_199 LDSPSHPRTASFLSKVLS--------------------------------
MUL_3658|M.ulcerans_Agy99 YDAPTNAFVMSFLGAVSALNG------------------ALVRPHDIRVG
Mb1270|M.bovis_AF2122/97 YERPANLFVAGFIGSPAMNFFPARLTAIGLTLPFGEVTLAPEVQGVIAAH
Rv1238|M.tuberculosis_H37Rv YERPANLFVAGFIGSPAMNFFPARLTAIGLTLPFGEVTLAPEVQGVIAAH
MAV_1377|M.avium_104 YENPANLFVAGFIGSPAMNFFPATLTPIGLKVPFGEVMLTPEVQQVIAEH
MLBr_01089|M.leprae_Br4923 YERPVNLFVAGFIGSPVMNFFPAKLTPNGLTLPLGELNLSRDVQAMIGQH
MMAR_1901|M.marinum_M YDNPVNTFVATFIGAPAMNLIEAPVAD-------GVARLSGLVIPVPRG-
Mflv_4055|M.gilvum_PYR-GCK LSDTALLAAHRLELPWGFTVGLR---------------------------
Mvan_2294|M.vanbaalenii_PYR-1 LSDADLLAAHRLELPWGFAVGSG---------------------------
MSMEG_2610|M.smegmatis_MC2_155 LGDAHLLAAHRLELPWGFAVPERSRN------------------------
TH_2525|M.thermoresistible__bu LCDEALLTXXXTGAAFNLSSVN----------------------------
MAB_4237c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_3658|M.ulcerans_Agy99 RTPEMAVAAADGTGESTGVTRAVVDRIVVLGFEVRVELTSAATGGAFTAQ
Mb1270|M.bovis_AF2122/97 PKPENVIVGVRPEHIQDAALIDAYQRIRALTFQVKVNLVESLGADKYLYF
Rv1238|M.tuberculosis_H37Rv PKPENVIVGVRPEHIQDAALIDAYQRIRALTFQVKVNLVESLGADKYLYF
MAV_1377|M.avium_104 PEPDNVIVGARPEHLSDAALIDGYQRIRALTFEVKVDMVESLGADKYVYF
MLBr_01089|M.leprae_Br4923 PVPDRVIVGVRPEHLTDAKLIDAHQHGTVLTFKVKVDLVESLGADKYIYF
MMAR_1901|M.marinum_M -AADRVLIGVRPE------SWDVVGGDAAAALGVRVEQVEELGFESFIYA
Mflv_4055|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2294|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2610|M.smegmatis_MC2_155 --------------------------------------------------
TH_2525|M.thermoresistible__bu --------------------------------------------------
MAB_4237c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_3658|M.ulcerans_Agy99 ITRGDAEALA-------------LQEGDTVYVRATRVPPIAGAAPAVPDD
Mb1270|M.bovis_AF2122/97 TTESPAVHSVQLDELAEVEGESALHENQFVARVPAESKVAIGQSVELAFD
Rv1238|M.tuberculosis_H37Rv TTESPAVHSVQLDELAEVEGESALHENQFVARVPAESKVAIGQSVELAFD
MAV_1377|M.avium_104 STAAWAAHSTQLDELAA---EADAHENQFVARVPAESKAAIGQTVELALD
MLBr_01089|M.leprae_Br4923 TTTGCDVYSAQLDELAS---ELEVRENQFVARVSAESKMAIGESIELAFG
MMAR_1901|M.marinum_M TPVAQDGWSS--------------RTRRIVIRSDRHTTVGAGDSLSIAPN
Mflv_4055|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2294|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2610|M.smegmatis_MC2_155 --------------------------------------------------
TH_2525|M.thermoresistible__bu --------------------------------------------------
MAB_4237c|M.abscessus_ATCC_199 -----------------------
MUL_3658|M.ulcerans_Agy99 GAGSTPRAAQIGVGGQ-------
Mb1270|M.bovis_AF2122/97 TARLAVFDADSGANLTIPHRA--
Rv1238|M.tuberculosis_H37Rv TARLAVFDADSGANLTIPHRA--
MAV_1377|M.avium_104 TTKLMVFDADSGVNLTVAPSDSP
MLBr_01089|M.leprae_Br4923 TAKIAVFDADSGVNLTVSAPTDA
MMAR_1901|M.marinum_M PQEVCFFDSRTESRIR-------
Mflv_4055|M.gilvum_PYR-GCK -----------------------
Mvan_2294|M.vanbaalenii_PYR-1 -----------------------
MSMEG_2610|M.smegmatis_MC2_155 -----------------------
TH_2525|M.thermoresistible__bu -----------------------