For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIVITGASGNVGTALLRRLIGTGDGHDLVGIVRRPPPAEGVYREVTWHAIDLADADAQDRLRPIFEGADA VVHLAWGFQPTRNTEYLTRVGVGGTRAVLGAAHAAAVGHLVHMSSVGAYAPGRYGDYVNEDWPTLGIRTS PYSRDKSRAESLLDEYLDEQGSAAIPVARLRPGFIVQRDAASGLMRYALPGYVPMPVVPLLPIVPLDRRL CIPLVHADDVAAALVAIVHRCAVGPFNVAAEPPVRRADVAEVLNARQIHVPSQILGTLVDLTWRARLQPI DRGWLDMAFSVPLLDCARAEKELGWQPTWSSVAALADVIDGVARQASTESPPLRPRSMLEQLRRDLTEGL MTTRHLP*
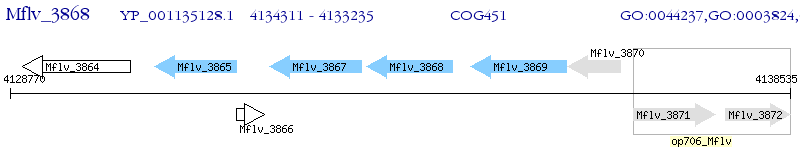
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3868 | - | - | 100% (358) | NAD-dependent epimerase/dehydratase |
| M. gilvum PYR-GCK | Mflv_1028 | - | 2e-28 | 31.33% (332) | NAD-dependent epimerase/dehydratase |
| M. gilvum PYR-GCK | Mflv_1108 | - | 1e-08 | 23.89% (339) | NAD-dependent epimerase/dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2073c | - | 8e-13 | 25.00% (320) | hypothetical protein Mb2073c |
| M. tuberculosis H37Rv | Rv2047c | - | 8e-13 | 25.00% (320) | hypothetical protein Rv2047c |
| M. leprae Br4923 | MLBr_02428 | - | 2e-06 | 22.57% (257) | putative glucose epimerase/dehydratase |
| M. abscessus ATCC 19977 | MAB_0718c | - | 7e-10 | 28.93% (121) | NAD-dependent epimerase/dehydratase |
| M. marinum M | MMAR_1810 | galE4 | 1e-26 | 33.23% (331) | UDP-glucose 4-epimerase, GalE4 |
| M. avium 104 | MAV_3754 | - | 4e-27 | 30.77% (338) | NAD dependent epimerase/dehydratase family protein |
| M. smegmatis MC2 155 | MSMEG_2247 | - | 4e-15 | 27.01% (348) | dihydrokaempferol 4-reductase |
| M. thermoresistible (build 8) | TH_4464 | - | 6e-24 | 31.53% (333) | PUTATIVE NAD-dependent epimerase/dehydratase |
| M. ulcerans Agy99 | MUL_2058 | - | 2e-26 | 32.93% (331) | hypothetical protein MUL_2058 |
| M. vanbaalenii PYR-1 | Mvan_5808 | - | 2e-27 | 30.56% (337) | NAD-dependent epimerase/dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1810|M.marinum_M -----------------MGVAVAVTGPTGDIGISAVTALEREPAVASIVG
MUL_2058|M.ulcerans_Agy99 -----------------MGVAVAVTGPTGDIGISAVTALEREPAVASIVG
MAV_3754|M.avium_104 -------------------MTVAVTGPTGEIGRSAVTALEREPGVDAIIG
TH_4464|M.thermoresistible__bu -----------------MALTVAVTGPTGEIGMAAVEALERDPAVGRIVG
Mvan_5808|M.vanbaalenii_PYR-1 -----------------MSLRVAVTGPTGEIGISTISALEAHPAVAEIIG
Mb2073c|M.bovis_AF2122/97 -------------------MRIAVTGASGVLGRGLTARLLSQG--HEVVG
Rv2047c|M.tuberculosis_H37Rv -------------------MRIAVTGASGVLGRGLTARLLSQG--HEVVG
Mflv_3868|M.gilvum_PYR-GCK -------------------MRIVITGASGNVGTALLRRLIGTGDGHDLVG
MAB_0718c|M.abscessus_ATCC_199 -------------------MKTAVTGAAGFIGTNLVNLLVDNG--HEVIA
MSMEG_2247|M.smegmatis_MC2_155 ---------------MDRRRKVLVMGASGNVGACVTRQLVERG--DDVRV
MLBr_02428|M.leprae_Br4923 MDSSSGQDGGGTGGVVPYPKVVLVTGACRFLGGYLTARLAQNPLINAVIA
: *. :* * :
MMAR_1810|M.marinum_M MARRPFDPAIYG-WSKTTYQRGNILDRDAVD---ALVAQADVVVHLAFIK
MUL_2058|M.ulcerans_Agy99 MARRPFDPAIYG-WSKTTYQRGNILDRDAVD---ALVAQADVVVHLAFIK
MAV_3754|M.avium_104 MARRPFSPSSRG-WQKTTYQQGDILDREAVD---AVVAQADVVIHLAFII
TH_4464|M.thermoresistible__bu MARRPFDAAAHG-WTKTEYRQGDILDREAVD---ALVAPADVVVHLAFII
Mvan_5808|M.vanbaalenii_PYR-1 MARRPFDTSEHG-WTKTTYRQGDILDRDAVE---ALVADTDVVVHLAFII
Mb2073c|M.bovis_AF2122/97 IARHRPDSWP----SSADFIAADIRDATAVE---SAMTGADVVAHCAWVR
Rv2047c|M.tuberculosis_H37Rv IARHRPDSWP----SSADFIAADIRDATAVE---SAMTGADVVAHCAWVR
Mflv_3868|M.gilvum_PYR-GCK IVRRPP-PAEGV-YREVTWHAIDLADADAQDRLRPIFEGADAVVHLAWGF
MAB_0718c|M.abscessus_ATCC_199 IDRVVPASPETR--EAVTWVSGDVLDPDSMT---KALDGVEVVYHLVAVI
MSMEG_2247|M.smegmatis_MC2_155 LLRRNSSTKGID-GLDVERHYGDIFDTGAVA---AAVADRDVVFYCVVDT
MLBr_02428|M.leprae_Br4923 VDAIAPSKDMLRRMGHAEFVRADIRNPFIAK--VIRNGEVDTVVHADAAS
: . :: : :.* :
MMAR_1810|M.marinum_M LGSR----AESAQVNLQGARNVFEATVAAR----RPRRLVYTSS------
MUL_2058|M.ulcerans_Agy99 LGSR----AESAQVNLQGARNVFEATVAAR----RPRRLVYTSS------
MAV_3754|M.avium_104 MGSR----DESARVNLQGTRNVFEATVAAG----RPRRLVYTSS------
TH_4464|M.thermoresistible__bu MGSR----AESERVNLTGTRNVFQAVVDHHARTGRPRRLVYTSS------
Mvan_5808|M.vanbaalenii_PYR-1 MGSR----EESARVNLEGTRNVFEAAVAG----PRVRRLVYTSS------
Mb2073c|M.bovis_AF2122/97 G--------RNDHINIDGTANVLKAMAETG-----TGRIVFTSSGHQPRV
Rv2047c|M.tuberculosis_H37Rv G--------RNDHINIDGTANVLKAMAETG-----TGRIVFTSSGHQPRV
Mflv_3868|M.gilvum_PYR-GCK QPTRNT--EYLTRVGVGGTRAVLGAAHAAA-----VGHLVHMSS------
MAB_0718c|M.abscessus_ATCC_199 TLKHED--ELCWRINTEGARTVAQAALTVG-----ARRMVHCSS------
MSMEG_2247|M.smegmatis_MC2_155 RAHLAAP-APLFRTNVEGLRNVLEVADHVD-----LHRFVFLST------
MLBr_02428|M.leprae_Br4923 YAPRSCGSAALKELNVMGAMQLFAACQKAP----TVRRVVLKST------
. . * : . :.* *:
MMAR_1810|M.marinum_M ----------------VAAYGYHSDNPAA--ITEDVPARGSPE------H
MUL_2058|M.ulcerans_Agy99 ----------------MAAYGYHSDNPAA--ITEDVPARGSPE------H
MAV_3754|M.avium_104 ----------------VAAYGYHSDNPVP--LTEDVPARGSAE------H
TH_4464|M.thermoresistible__bu ----------------VAAYGYHSDNPVP--ITESVAPRGSPE------H
Mvan_5808|M.vanbaalenii_PYR-1 ----------------VAAYGYHSDNPVP--ITEDVPPRGSPE------H
Mb2073c|M.bovis_AF2122/97 EQMLADCGLEWVAVRCALIFGRNVDNWVQRLFALPVLPAGYADRVVQVVH
Rv2047c|M.tuberculosis_H37Rv EQMLADCGLEWVAVRCALIFGRNVDNWVQRLFALPVLPAGYADRVVQVVH
Mflv_3868|M.gilvum_PYR-GCK -------------------VGAYAPGRYGDYVNEDWPTLGIRT------S
MAB_0718c|M.abscessus_ATCC_199 -------------------IDSYSNSVAT--IDEKSPRSAAAD-----LP
MSMEG_2247|M.smegmatis_MC2_155 -------------------IGTIAVGRNGEAVDEDTPFNWAGIG-----G
MLBr_02428|M.leprae_Br4923 ------------------SAVYGSGSRDPVMFTEDSSSWRSFR------E
. .
MMAR_1810|M.marinum_M CYSEQKAQSEAELAKITED-------------------------------
MUL_2058|M.ulcerans_Agy99 YYSEQKAQSEAELAKITED-------------------------------
MAV_3754|M.avium_104 YYSAQKAASEAMLAEITKD-------------------------------
TH_4464|M.thermoresistible__bu YYSAQKAACEALLAEITDG-------------------------------
Mvan_5808|M.vanbaalenii_PYR-1 YYSEQKAACEALLADVTAG-------------------------------
Mb2073c|M.bovis_AF2122/97 SDDAQRLLVRALLDTVIDSGPVNLAAPGELTFRRIAAALGRPMVPIGSPV
Rv2047c|M.tuberculosis_H37Rv SDDAQRLLVRALLDTVIDSGPVNLAAPGELTFRRIAAALGRPMVPIGSPV
Mflv_3868|M.gilvum_PYR-GCK PYSRDKSRAESLLDEYLDE-------------------------------
MAB_0718c|M.abscessus_ATCC_199 VYQRSKWGGEVAVRETIAS-------------------------------
MSMEG_2247|M.smegmatis_MC2_155 PYIESRRKAEELVLSYAAER------------------------------
MLBr_02428|M.leprae_Br4923 GFAKDSLDIEGYARGLGRRR------------------------------
. .
MMAR_1810|M.marinum_M ----------------SLLQVFVLRP------------------------
MUL_2058|M.ulcerans_Agy99 ----------------SLLQVFVLRP------------------------
MAV_3754|M.avium_104 ----------------SPLEVFVLRP------------------------
TH_4464|M.thermoresistible__bu ----------------SGVEVYVLRP------------------------
Mvan_5808|M.vanbaalenii_PYR-1 ----------------SALEVYVLRP------------------------
Mb2073c|M.bovis_AF2122/97 LRRVTSFAELELLHSAPLMDVTLLRDRWGFQPAWNAEECLEDFTLAVRGR
Rv2047c|M.tuberculosis_H37Rv LRRVTSFAELELLHSAPLMDVTLLRDRWGFQPAWNAEECLEDFTLAVRGR
Mflv_3868|M.gilvum_PYR-GCK -------------QGSAAIPVARLRPG-----------------------
MAB_0718c|M.abscessus_ATCC_199 -----------------GLDAVICNP------------------------
MSMEG_2247|M.smegmatis_MC2_155 -----------------GFPAVVMNVS-----------------------
MLBr_02428|M.leprae_Br4923 ----------------SDIAVTILRLA-----------------------
. . .
MMAR_1810|M.marinum_M --------------------------------CIVAGPH-----------
MUL_2058|M.ulcerans_Agy99 --------------------------------CIVAGPH-----------
MAV_3754|M.avium_104 --------------------------------CIVAGPG-----------
TH_4464|M.thermoresistible__bu --------------------------------CIVAGPR-----------
Mvan_5808|M.vanbaalenii_PYR-1 --------------------------------CIVAGPK-----------
Mb2073c|M.bovis_AF2122/97 IGLGKRTFSLPWRLANIQDLPAVDSPADDGVAPRLAGPEGANGEFDTPID
Rv2047c|M.tuberculosis_H37Rv IGLGKRTFSLPWRLANIQDLPAVDSPADDGVAPRLAGPEGANGEFDTPID
Mflv_3868|M.gilvum_PYR-GCK --------------------------------FIVQRDA-----------
MAB_0718c|M.abscessus_ATCC_199 --------------------------------TGVYGPVD----------
MSMEG_2247|M.smegmatis_MC2_155 ---------------------------------NPYGPPD----------
MLBr_02428|M.leprae_Br4923 ---------------------------------DMIGPAM----------
MMAR_1810|M.marinum_M -------AYSLAEAMPCRQVP-----------------------------
MUL_2058|M.ulcerans_Agy99 -------AYSLAEAMPCRQVP-----------------------------
MAV_3754|M.avium_104 -------ATALADAMPWNQLP-----------------------------
TH_4464|M.thermoresistible__bu -------APALADAMPWNQLP-----------------------------
Mvan_5808|M.vanbaalenii_PYR-1 -------ATALADAMPWNQLP-----------------------------
Mb2073c|M.bovis_AF2122/97 PRFPTYLATNLSEALPGPFSPSSASVTVRGLRAGGVGIAERLRPSGVIQR
Rv2047c|M.tuberculosis_H37Rv PRFPTYLATNLSEALPGPFSPSSASVTVRGLRAGGVGIAERLRPSGVIQR
Mflv_3868|M.gilvum_PYR-GCK -------ASGLMRYALPGYVP-----------------------------
MAB_0718c|M.abscessus_ATCC_199 -------LPNLSRINQLLFDS-----------------------------
MSMEG_2247|M.smegmatis_MC2_155 ------WQPRQGALVAMAAFG-----------------------------
MLBr_02428|M.leprae_Br4923 -------DTTLSRYLAGPLVP-----------------------------
MMAR_1810|M.marinum_M --------------------------------------------------
MUL_2058|M.ulcerans_Agy99 --------------------------------------------------
MAV_3754|M.avium_104 --------------------------------------------------
TH_4464|M.thermoresistible__bu --------------------------------------------------
Mvan_5808|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 EIAMRTVAVFAHRLYGAITSAHFMAATVPFAKPATIVSNSGFFGPSMASL
Rv2047c|M.tuberculosis_H37Rv EIAMRTVAVFAHRLYGAITSAHFMAATVPFAKPATIVSNSGFFGPSMASL
Mflv_3868|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0718c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_2247|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02428|M.leprae_Br4923 --------------------------------------------------
MMAR_1810|M.marinum_M --------------------------------------------------
MUL_2058|M.ulcerans_Agy99 --------------------------------------------------
MAV_3754|M.avium_104 --------------------------------------------------
TH_4464|M.thermoresistible__bu --------------------------------------------------
Mvan_5808|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 PIFGAQRPPSESSRARRWLRTLRNIGVFGVNLVGLSAGSPRDTDAYVADV
Rv2047c|M.tuberculosis_H37Rv PIFGAQRPPSESSRARRWLRTLRNIGVFGVNLVGLSAGSPRDTDAYVADV
Mflv_3868|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0718c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_2247|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02428|M.leprae_Br4923 --------------------------------------------------
MMAR_1810|M.marinum_M --------------------------------------------------
MUL_2058|M.ulcerans_Agy99 --------------------------------------------------
MAV_3754|M.avium_104 --------------------------------------------------
TH_4464|M.thermoresistible__bu --------------------------------------------------
Mvan_5808|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 DRLERLAFDNLATHDDRRLLSLILLARDHVVHGWVLASGSFMLCAAFNVL
Rv2047c|M.tuberculosis_H37Rv DRLERLAFDNLATHDDRRLLSLILLARDHVVHGWVLASGSFMLCAAFNVL
Mflv_3868|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0718c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_2247|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02428|M.leprae_Br4923 --------------------------------------------------
MMAR_1810|M.marinum_M -----------------------APVRALSR---VVPMLKPIVPDPG---
MUL_2058|M.ulcerans_Agy99 -----------------------APVRALSR---VVPMLKPIVPDPG---
MAV_3754|M.avium_104 -----------------------GPLRAIVKAVQAIPVLKPVVPDPG---
TH_4464|M.thermoresistible__bu -----------------------KPVKRITQ---ALPLLRPPFPDPG---
Mvan_5808|M.vanbaalenii_PYR-1 -----------------------GPVRRLAH---ALPLLKPPFPDPG---
Mb2073c|M.bovis_AF2122/97 LRGLCGRDTAPAAGPELVSARSVEAVQRLVAAARRDPVVIRLLAEPGERL
Rv2047c|M.tuberculosis_H37Rv LRGLCGRDTAPAAGPELVSARSVEAVQRLVAAARRDPVVIRLLAEPGERL
Mflv_3868|M.gilvum_PYR-GCK --------------------------------MPVVPLLPIVPLDRR---
MAB_0718c|M.abscessus_ATCC_199 -----------------------ARGRLPGMVNSQYDLVDARDVAAG---
MSMEG_2247|M.smegmatis_MC2_155 --------------------------KLPVYIRGVGAEVVGIDDAAR---
MLBr_02428|M.leprae_Br4923 ---------------------------------------TMCGRDAR---
MMAR_1810|M.marinum_M ---------------VPVQLVHHDDVAAAIALAATAP-VPAGAYNIAADG
MUL_2058|M.ulcerans_Agy99 ---------------VPVQLVHHDDVAAAIALAATAP-VPAGAYNIAADG
MAV_3754|M.avium_104 ---------------YPLQLVHHDDVASAIALAATAP-APPGAYNIAGDG
TH_4464|M.thermoresistible__bu ---------------TPLQLVHHDDVAEAIRLAATTTSAPPGAYNIAADG
Mvan_5808|M.vanbaalenii_PYR-1 ---------------TPVQLVHHDDVASAIVAAVTTTSAQPGAYNIAGDG
Mb2073c|M.bovis_AF2122/97 DKLAVEAPEFHSAVLAELTLIGHRGPAEVEMAATSYADNPELLVRMVAKT
Rv2047c|M.tuberculosis_H37Rv DKLAVEAPEFHSAVLAELTLIGHRGPAEVEMAATSYADNPELLVRMVAKT
Mflv_3868|M.gilvum_PYR-GCK ---------------LCIPLVHADDVAAALVAIVHRC--AVGPFNVAAEP
MAB_0718c|M.abscessus_ATCC_199 -----------------LYLAGEKGRTGENYLLGGHMGSLLQVCRLAAR-
MSMEG_2247|M.smegmatis_MC2_155 ----------------ALILAAEHGRIGERYIVSERYMSQQEMLTVAAEA
MLBr_02428|M.leprae_Br4923 -----------------LQLLHEQDALGALECAAMTG--KAGTFNIGASG
: * . : .
MMAR_1810|M.marinum_M VVTITDMARALGGRPVRVPAAAASVASAAISR------------------
MUL_2058|M.ulcerans_Agy99 VVTITDMARALGGRPVRVPAAAASVASAAISR------------------
MAV_3754|M.avium_104 VVTVADVAKALGGRPVRVPAVAATAASAAISK------------------
TH_4464|M.thermoresistible__bu VLSMSQVASALGARPIRVPRAAMTATSELVAR------------------
Mvan_5808|M.vanbaalenii_PYR-1 VLSMSAVGEALGARPVRVPHGAATATSEVIAR------------------
Mb2073c|M.bovis_AF2122/97 LRAVPAPQPPTPVIPLRAKPVALLAARQLRDREVRRDRMVRAIWVLRALL
Rv2047c|M.tuberculosis_H37Rv LRAVPAPQPPTPVIPLRAKPVALLAARQLRDREVRRDRMVRAIWVLRALL
Mflv_3868|M.gilvum_PYR-GCK PVRRADVAEVLNARQIHVPSQILGTLVDLTWR------------------
MAB_0718c|M.abscessus_ATCC_199 RAGKRGPRFAVPMGLLNAAIPVIEPIGKLLKN------------------
MSMEG_2247|M.smegmatis_MC2_155 VGARP-PRIGIPMAAVYAFGSLAGLSNRLFRT------------------
MLBr_02428|M.leprae_Br4923 IMMLSQAIRRAGRIAVPIPSFGVWALDLLRWVN-----------------
:
MMAR_1810|M.marinum_M -------------------LPLVPPLLEWLHAVRQPVVMDTAKAKRELG-
MUL_2058|M.ulcerans_Agy99 -------------------LPLVPPLLEWLHAVRQPVVMDTAKAKRELG-
MAV_3754|M.avium_104 -------------------APLVPSMLEWLHTARTSMVMDTTKAKTQLG-
TH_4464|M.thermoresistible__bu -------------------LPFIPSTLEWLHVGRTSVVMDVDKAKRLLG-
Mvan_5808|M.vanbaalenii_PYR-1 -------------------LPFVPSILEWLHAGRTSVVMDTTKARDQLG-
Mb2073c|M.bovis_AF2122/97 REYGRRLTEAGVFDTPDDVFYLLVDEIDALPADVSGLVARRRAEQRRLAG
Rv2047c|M.tuberculosis_H37Rv REYGRRLTEAGVFDTPDDVFYLLVDEIDALPADVSGLVARRRAEQRRLAG
Mflv_3868|M.gilvum_PYR-GCK -------------------ARLQPIDRGWLDMAFSVPLLDCARAEKELG-
MAB_0718c|M.abscessus_ATCC_199 ----------------------DALSRAAFNKLTVSPAVDITKARTELG-
MSMEG_2247|M.smegmatis_MC2_155 ---------------------DFPINLTAARLMWWTSPADHSKATRELG-
MLBr_02428|M.leprae_Br4923 --------------------RYTEITRDQFEYLSYGRVMDTTRMGSELG-
*.
MMAR_1810|M.marinum_M ----------WSPTHSSAETLSALAAAI----------------------
MUL_2058|M.ulcerans_Agy99 ----------WSPTHSSAETLSALAAAI----------------------
MAV_3754|M.avium_104 ----------WRPVHTSAQALAALASAV----------------------
TH_4464|M.thermoresistible__bu ----------WTPTHTAADTLAALAARPT---------------------
Mvan_5808|M.vanbaalenii_PYR-1 ----------WRPNFSAAETLSALASSIGND-------------------
Mb2073c|M.bovis_AF2122/97 IVPPTVFSGSWEPSPSSAAALAAGDTLRGVGVCGGRVRGRVRIVRPETID
Rv2047c|M.tuberculosis_H37Rv IVPPTVFSGSWEPSPSSAAALAAGDTLRGVGVCGGRVRGRVRIVRPETID
Mflv_3868|M.gilvum_PYR-GCK ----------WQPTWSSVAALADVIDGVARQASTESPPLRPRSMLEQLRR
MAB_0718c|M.abscessus_ATCC_199 ----------YEPRSTETTVNEFVDFLLGSGRL-----------------
MSMEG_2247|M.smegmatis_MC2_155 ----------WKPAPTADAIARAAQFYVERKNNNEKVISL----------
MLBr_02428|M.leprae_Br4923 ----------YHPKWTTAGAFDDYVRGRGLTPIIDLHRVRYFEGRAIALA
: * :
MMAR_1810|M.marinum_M --------------------------------------------------
MUL_2058|M.ulcerans_Agy99 --------------------------------------------------
MAV_3754|M.avium_104 --------------------------------------------------
TH_4464|M.thermoresistible__bu --------------------------------------------------
Mvan_5808|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 DLQPGEILVAEVTDVGYTAAFCYAAAVVTELGGPMSHAAVVAREFGFPCV
Rv2047c|M.tuberculosis_H37Rv DLQPGEILVAEVTDVGYTAAFCYAAAVVTELGGPMSHAAVVAREFGFPCV
Mflv_3868|M.gilvum_PYR-GCK DLTEGLMTTRHLP-------------------------------------
MAB_0718c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_2247|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02428|M.leprae_Br4923 QRWSSRN-------------------------------------------
MMAR_1810|M.marinum_M ---------------------------------------------
MUL_2058|M.ulcerans_Agy99 ---------------------------------------------
MAV_3754|M.avium_104 ---------------------------------------------
TH_4464|M.thermoresistible__bu ---------------------------------------------
Mvan_5808|M.vanbaalenii_PYR-1 ---------------------------------------------
Mb2073c|M.bovis_AF2122/97 VDAQGATRFLPPGALVEVDGATGEIHVVELASEDGPALPGSDLSR
Rv2047c|M.tuberculosis_H37Rv VDAQGATRFLPPGALVEVDGATGEIHVVELASEDGPALPGSDLSR
Mflv_3868|M.gilvum_PYR-GCK ---------------------------------------------
MAB_0718c|M.abscessus_ATCC_199 ---------------------------------------------
MSMEG_2247|M.smegmatis_MC2_155 ---------------------------------------------
MLBr_02428|M.leprae_Br4923 ---------------------------------------------