For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVAVTGPTGEIGRSAVTALEREPGVDAIIGMARRPFSPSSRGWQKTTYQQGDILDREAVDAVVAQADVVI HLAFIIMGSRDESARVNLQGTRNVFEATVAAGRPRRLVYTSSVAAYGYHSDNPVPLTEDVPARGSAEHYY SAQKAASEAMLAEITKDSPLEVFVLRPCIVAGPGATALADAMPWNQLPGPLRAIVKAVQAIPVLKPVVPD PGYPLQLVHHDDVASAIALAATAPAPPGAYNIAGDGVVTVADVAKALGGRPVRVPAVAATAASAAISKAP LVPSMLEWLHTARTSMVMDTTKAKTQLGWRPVHTSAQALAALASAV*
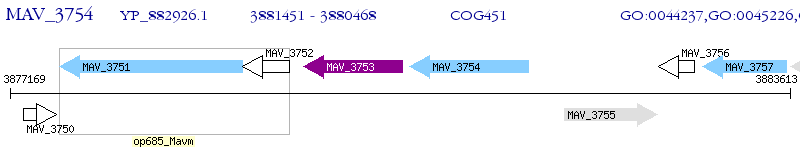
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3754 | - | - | 100% (327) | NAD dependent epimerase/dehydratase family protein |
| M. avium 104 | MAV_2451 | - | 1e-17 | 29.66% (327) | hypothetical protein MAV_2451 |
| M. avium 104 | MAV_4649 | - | 5e-16 | 25.30% (328) | NAD dependent epimerase/dehydratase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0513 | galE2 | 9e-16 | 24.38% (324) | UDP-glucose 4-epimerase |
| M. gilvum PYR-GCK | Mflv_1028 | - | 1e-131 | 71.08% (325) | NAD-dependent epimerase/dehydratase |
| M. tuberculosis H37Rv | Rv0501 | galE2 | 9e-16 | 24.38% (324) | UDP-glucose 4-epimerase |
| M. leprae Br4923 | MLBr_02428 | - | 4e-13 | 24.46% (327) | putative glucose epimerase/dehydratase |
| M. abscessus ATCC 19977 | MAB_4003c | - | 4e-17 | 26.44% (329) | putative UDP-glucose 4-epimerase GalE1 |
| M. marinum M | MMAR_1810 | galE4 | 1e-137 | 73.70% (327) | UDP-glucose 4-epimerase, GalE4 |
| M. smegmatis MC2 155 | MSMEG_0946 | - | 3e-15 | 25.46% (326) | NAD dependent epimerase/dehydratase family protein |
| M. thermoresistible (build 8) | TH_4464 | - | 1e-128 | 70.00% (330) | PUTATIVE NAD-dependent epimerase/dehydratase |
| M. ulcerans Agy99 | MUL_2058 | - | 1e-137 | 73.70% (327) | hypothetical protein MUL_2058 |
| M. vanbaalenii PYR-1 | Mvan_5808 | - | 1e-131 | 71.47% (326) | NAD-dependent epimerase/dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1810|M.marinum_M ---------------------MGVAVAVTGPTGDIGISAVTALEREPAVA
MUL_2058|M.ulcerans_Agy99 ---------------------MGVAVAVTGPTGDIGISAVTALEREPAVA
MAV_3754|M.avium_104 -----------------------MTVAVTGPTGEIGRSAVTALEREPGVD
Mflv_1028|M.gilvum_PYR-GCK ---------------------MSLRVAVTGPTGEIGISTISALEALPEVT
Mvan_5808|M.vanbaalenii_PYR-1 ---------------------MSLRVAVTGPTGEIGISTISALEAHPAVA
TH_4464|M.thermoresistible__bu ---------------------MALTVAVTGPTGEIGMAAVEALERDPAVG
Mb0513|M.bovis_AF2122/97 MSSSNGRGGAGGVGGSSEHPQYPKVVLVTGACRFLGGYLTARLAQNPLIN
Rv0501|M.tuberculosis_H37Rv MSSSNGRGGAGGVGGSSEHPQYPKVVLVTGACRFLGGYLTARLAQNPLIN
MLBr_02428|M.leprae_Br4923 MDSSSGQDG-GGTGG---VVPYPKVVLVTGACRFLGGYLTARLAQNPLIN
MSMEG_0946|M.smegmatis_MC2_155 ------------------------MVLVTGACRFLGGYLTARLAQNPSID
MAB_4003c|M.abscessus_ATCC_199 MSAGESN------------LHYPRVVLVTGASRFLGGYLAARLVQNPMIN
* ***. :* * * :
MMAR_1810|M.marinum_M SIVGMARR-PFDPAIYGWSKTTYQRGNILDRDAVDALVAQ-ADVVVHLAF
MUL_2058|M.ulcerans_Agy99 SIVGMARR-PFDPAIYGWSKTTYQRGNILDRDAVDALVAQ-ADVVVHLAF
MAV_3754|M.avium_104 AIIGMARR-PFSPSSRGWQKTTYQQGDILDREAVDAVVAQ-ADVVIHLAF
Mflv_1028|M.gilvum_PYR-GCK EIVGMARR-PFDPAAQGWTKTLYRQGDILDRDAVERLVED-ADVVVHLAF
Mvan_5808|M.vanbaalenii_PYR-1 EIIGMARR-PFDTSEHGWTKTTYRQGDILDRDAVEALVAD-TDVVVHLAF
TH_4464|M.thermoresistible__bu RIVGMARR-PFDAAAHGWTKTEYRQGDILDREAVDALVAP-ADVVVHLAF
Mb0513|M.bovis_AF2122/97 RVIAVDAIAPSKDMLRRMGRAEFVRADIRNPFIAKVIRNGEVDTVVHAAA
Rv0501|M.tuberculosis_H37Rv RVIAVDAIAPSKDMLRRMGRAEFVRADIRNPFIAKVIRNGEVDTVVHAAA
MLBr_02428|M.leprae_Br4923 AVIAVDAIAPSKDMLRRMGHAEFVRADIRNPFIAKVIRNGEVDTVVHADA
MSMEG_0946|M.smegmatis_MC2_155 HVIAVDAITPSKDLLRRMGRAEFVRADIRNPFIAKVIRNGNVDTVVHAAA
MAB_4003c|M.abscessus_ATCC_199 RVIAVDAVAPSKDLLRRMGRAEFVRADIRNPFIAKVIRNGEVDTVVHTAS
::.: * . :: : :.:* : .. : .*.*:*
MMAR_1810|M.marinum_M IKLGSRAES----AQVNLQGARNVFEATVAAR----RPRRLVYTSSVAAY
MUL_2058|M.ulcerans_Agy99 IKLGSRAES----AQVNLQGARNVFEATVAAR----RPRRLVYTSSMAAY
MAV_3754|M.avium_104 IIMGSRDES----ARVNLQGTRNVFEATVAAG----RPRRLVYTSSVAAY
Mflv_1028|M.gilvum_PYR-GCK IIMGSREES----ARVNLAGTRNVFEAVVAA----ARPRRLVYTSSVAAY
Mvan_5808|M.vanbaalenii_PYR-1 IIMGSREES----ARVNLEGTRNVFEAAVAG----PRVRRLVYTSSVAAY
TH_4464|M.thermoresistible__bu IIMGSRAES----ERVNLTGTRNVFQAVVDHHARTGRPRRLVYTSSVAAY
Mb0513|M.bovis_AF2122/97 ASYAPRSGGSAALKELNVMGAMQLFAACQKAP----SVRRVVLKSTSEVY
Rv0501|M.tuberculosis_H37Rv ASYAPRSGGSAALKELNVMGAMQLFAACQKAP----SVRRVVLKSTSEVY
MLBr_02428|M.leprae_Br4923 ASYAPRSCGSAALKELNVMGAMQLFAACQKAP----TVRRVVLKSTSAVY
MSMEG_0946|M.smegmatis_MC2_155 ASYAPRSGGRATLKELNVMGAIQLFAACQKTP----SVRRVILKSTSEVY
MAB_4003c|M.abscessus_ATCC_199 ASYSPRSGGRAALKELNVMGAMQLFAACQKAP----TVQRVVVKSTAQVY
..* . .:*: *: ::* * :*:: .*: .*
MMAR_1810|M.marinum_M GYHSDNPAAITEDVPARGSPEHCYSEQKAQSEAELAKITED-SLLQVFVL
MUL_2058|M.ulcerans_Agy99 GYHSDNPAAITEDVPARGSPEHYYSEQKAQSEAELAKITED-SLLQVFVL
MAV_3754|M.avium_104 GYHSDNPVPLTEDVPARGSAEHYYSAQKAASEAMLAEITKD-SPLEVFVL
Mflv_1028|M.gilvum_PYR-GCK GYHADNPVPITEDVPVRGSAEHYYSEQKAACEAALREVTAG-SDLEVYVL
Mvan_5808|M.vanbaalenii_PYR-1 GYHSDNPVPITEDVPPRGSPEHYYSEQKAACEALLADVTAG-SALEVYVL
TH_4464|M.thermoresistible__bu GYHSDNPVPITESVAPRGSPEHYYSAQKAACEALLAEITDG-SGVEVYVL
Mb0513|M.bovis_AF2122/97 GSSPHDPVMFTEDSSSRRPFSQGFPKDSLDIEGYVRALGRRRPDIAVTIL
Rv0501|M.tuberculosis_H37Rv GSSPHDPVMFTEDSSSRRPFSQGFPKDSLDIEGYVRALGRRRPDIAVTIL
MLBr_02428|M.leprae_Br4923 GSGSRDPVMFTEDSSSWRSFREGFAKDSLDIEGYARGLGRRRSDIAVTIL
MSMEG_0946|M.smegmatis_MC2_155 GSSSRDPVMFTEESSARRPPRDGFARDSIDIEGYARGLARRRPDIAVTIL
MAB_4003c|M.abscessus_ATCC_199 GASARDPVMFTEEMSARRPPADGFARDSIDIESYARGLGRRRPDVAVTIL
* . :*. :**. . . . :. :. *. : . : * :*
MMAR_1810|M.marinum_M RPCIVAGPHAYSLAEAMPCRQVPAPVRALSR---VVPMLKPIVPDPGVPV
MUL_2058|M.ulcerans_Agy99 RPCIVAGPHAYSLAEAMPCRQVPAPVRALSR---VVPMLKPIVPDPGVPV
MAV_3754|M.avium_104 RPCIVAGPGATALADAMPWNQLPGPLRAIVKAVQAIPVLKPVVPDPGYPL
Mflv_1028|M.gilvum_PYR-GCK RPCIVAGPKATALADAMPWNQLPMPVQRVAQ---ALPLLKPPFPDPGTPL
Mvan_5808|M.vanbaalenii_PYR-1 RPCIVAGPKATALADAMPWNQLPGPVRRLAH---ALPLLKPPFPDPGTPV
TH_4464|M.thermoresistible__bu RPCIVAGPRAPALADAMPWNQLPKPVKRITQ---ALPLLRPPFPDPGTPL
Mb0513|M.bovis_AF2122/97 RLANMIGP----AMDTTLSRYLAGP-------------LVPTIFGRDARL
Rv0501|M.tuberculosis_H37Rv RLANMIGP----AMDTTLSRYLAGP-------------LVPTIFGRDARL
MLBr_02428|M.leprae_Br4923 RLADMIGP----AMDTTLSRYLAGP-------------LVPTMCGRDARL
MSMEG_0946|M.smegmatis_MC2_155 RLANMIGP----AMDTALSRYLAGP-------------VVPCVVGHDARL
MAB_4003c|M.abscessus_ATCC_199 RLANLIGP----GMDTALARYLAGP-------------VVPTMLGRDARL
* . : ** :: . :. * : * . . . :
MMAR_1810|M.marinum_M QLVHHDDVAAAIALAATAP-VPAGAYNIAADGVVTITDMARALGGRPVRV
MUL_2058|M.ulcerans_Agy99 QLVHHDDVAAAIALAATAP-VPAGAYNIAADGVVTITDMARALGGRPVRV
MAV_3754|M.avium_104 QLVHHDDVASAIALAATAP-APPGAYNIAGDGVVTVADVAKALGGRPVRV
Mflv_1028|M.gilvum_PYR-GCK QLVHHDDVAAAIALAVTTTTAPPGAYNIAGDGVLSMSALGEALGARPVPV
Mvan_5808|M.vanbaalenii_PYR-1 QLVHHDDVASAIVAAVTTTSAQPGAYNIAGDGVLSMSAVGEALGARPVRV
TH_4464|M.thermoresistible__bu QLVHHDDVAEAIRLAATTTSAPPGAYNIAADGVLSMSQVASALGARPIRV
Mb0513|M.bovis_AF2122/97 QLLHEQDALGALERAAMAG--KAGTFNIGADGILMLSQAIRRAGRIPVPV
Rv0501|M.tuberculosis_H37Rv QLLHEQDALGALERAAMAG--KAGTFNIGADGILMLSQAIRRAGRIPVPV
MLBr_02428|M.leprae_Br4923 QLLHEQDALGALECAAMTG--KAGTFNIGASGIMMLSQAIRRAGRIAVPI
MSMEG_0946|M.smegmatis_MC2_155 QLLHEQDALGALEHATVAG--KAGTFNIGADGIIMMSQALRRSGRIRLPV
MAB_4003c|M.abscessus_ATCC_199 QLLHEQDALGALERATMAG--KAGTFNIAADGMMMMSQAVRRSGQLGIPV
**:*.:*. *: *. : .*::**...*:: :: * : :
MMAR_1810|M.marinum_M PAAAASVASAAISRLPLVPPLLEWLHAVRQPVVMDTAKAKRELGWSPTHS
MUL_2058|M.ulcerans_Agy99 PAAAASVASAAISRLPLVPPLLEWLHAVRQPVVMDTAKAKRELGWSPTHS
MAV_3754|M.avium_104 PAVAATAASAAISKAPLVPSMLEWLHTARTSMVMDTTKAKTQLGWRPVHT
Mflv_1028|M.gilvum_PYR-GCK PRAAAVAASEVIARLPFVPSSLEWLHAGRTSVVMDTGKARDQLGWRPKYS
Mvan_5808|M.vanbaalenii_PYR-1 PHGAATATSEVIARLPFVPSILEWLHAGRTSVVMDTTKARDQLGWRPNFS
TH_4464|M.thermoresistible__bu PRAAMTATSELVARLPFIPSTLEWLHVGRTSVVMDVDKAKRLLGWTPTHT
Mb0513|M.bovis_AF2122/97 PGFGVWALDSLRRANHYTELNREQFAYLSYGRVMDTTRMRVELGYQPKWT
Rv0501|M.tuberculosis_H37Rv PGFGVWALDSLRRANHYTELNREQFAYLSYGRVMDTTRMRVELGYQPKWT
MLBr_02428|M.leprae_Br4923 PSFGVWALDLLRWVNRYTEITRDQFEYLSYGRVMDTTRMGSELGYHPKWT
MSMEG_0946|M.smegmatis_MC2_155 PRSALAAVDSLNRATRYSEVDREQLNYLSYGRVMDTTRMRRDLGYSPKWT
MAB_4003c|M.abscessus_ATCC_199 PSFAVAAIASFTKGTRYTELSSEQRDWLAYGRAMDNTRMKTELGYQPKWT
* . . : .** : **: * :
MMAR_1810|M.marinum_M SAETLSALAAAI-------------------------------------
MUL_2058|M.ulcerans_Agy99 SAETLSALAAAI-------------------------------------
MAV_3754|M.avium_104 SAQALAALASAV-------------------------------------
Mflv_1028|M.gilvum_PYR-GCK AAETLSALAAARGHGG---------------------------------
Mvan_5808|M.vanbaalenii_PYR-1 AAETLSALASSIGND----------------------------------
TH_4464|M.thermoresistible__bu AADTLAALAARPT------------------------------------
Mb0513|M.bovis_AF2122/97 TVEAFDDYFRGRGLTPIIDPHRVRSWEGRAVGLAQRWGSRNPIPWSGLR
Rv0501|M.tuberculosis_H37Rv TVEAFDDYFRGRGLTPIIDPHRVRSWEGRAVGLAQRWGSRNPIPWSGLR
MLBr_02428|M.leprae_Br4923 TAGAFDDYVRGRGLTPIIDLHRVRYFEGRAIALAQRWSSRN--------
MSMEG_0946|M.smegmatis_MC2_155 TGEAFDDYVRGRGLTPIIDPRWVGSLESRAVALAQRWGR----------
MAB_4003c|M.abscessus_ATCC_199 TIGAFGDYVRGRGITPVIEPEWVRSLGDRAVAFAQKISS----------
: ::