For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRLGRPPAGLLLPAALVAACTLVPLVYLFERAFERGLPFVVDELLQPRTAAMVGRSLLLVSVVTAACVVL GVGLAVLVTRTDLRGRRVLAVALTLPLAMPSYLLAYLWVSAFPTAAGFWGAALVLTLVSYPLVLLTTMAA LARVDPAQEEVARSLGLGGVATLFRVTLRQTRAAIAAGALLVALYVLSDFGAVAAMRYEAFTWVIYGAYR SGFNPSRAAVLSLVLIVFAVLLVVGEHRARGVAAAGRIGGGAPRPAPVSRLGRWRWPALAVPAVVLTAAL VVPGIELMDWLLAAGMQWNVGEWLGALGSTVWLSAAAALVSTAAALPLGVLAARHRDRVTRALEGASFLA HGLPSIVIAISMVSLGVLLLRPIYQREPLLILAYAVLFVPMAIGSIRASVQATPVRLEEVARSLGRPPVR AFATVTARVALPGIAAGAALVLLTCMKELPVTLLLHPTGTDTLATRLWGYSSISDYAAAAPYAAALLLFA AVPTAVLGLWSTRSAEVRVD*
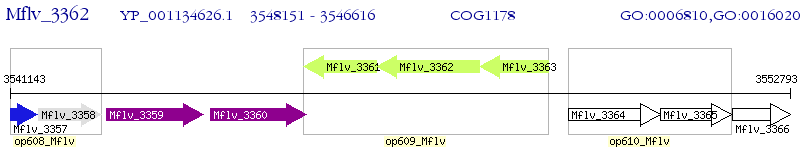
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3362 | - | - | 100% (511) | binding-protein-dependent transport systems inner membrane component |
| M. gilvum PYR-GCK | Mflv_2857 | - | 4e-94 | 38.45% (502) | binding-protein-dependent transport systems inner membrane |
| M. gilvum PYR-GCK | Mflv_2680 | - | 1e-16 | 26.14% (241) | sulfate ABC transporter, inner membrane subunit CysW |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2420c | cysW | 4e-17 | 27.53% (247) | sulfate-transport integral membrane protein ABC transporter |
| M. tuberculosis H37Rv | Rv2398c | cysW | 7e-16 | 27.13% (247) | sulfate-transport integral membrane protein ABC transporter |
| M. leprae Br4923 | MLBr_02093 | pstA1 | 7e-08 | 26.40% (197) | membrane-bound component of phosphate transport |
| M. abscessus ATCC 19977 | MAB_1654 | - | 1e-16 | 26.92% (260) | sulfate ABC transporter, permease protein CysW |
| M. marinum M | MMAR_3716 | cysW | 1e-16 | 28.03% (239) | sulfate-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_1784 | cysW | 1e-13 | 26.14% (241) | sulfate ABC transporter, permease protein CysW |
| M. smegmatis MC2 155 | MSMEG_3635 | - | 0.0 | 77.09% (502) | iron(III)-transport system permease protein SfuB |
| M. thermoresistible (build 8) | TH_0284 | - | 1e-106 | 80.00% (250) | iron(III)-transport system permease protein SfuB |
| M. ulcerans Agy99 | MUL_3659 | cysW | 6e-17 | 28.03% (239) | sulfate-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_3092 | - | 0.0 | 86.96% (506) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3362|M.gilvum_PYR-GCK ------------------MSRLGRPPAGLLLPAALVAACTLVPLVYLFER
Mvan_3092|M.vanbaalenii_PYR-1 ---------------MPVITARSRPPALLLLPAALVAAGTLIPLVYLVER
MSMEG_3635|M.smegmatis_MC2_155 ---------------------------MLVAAGAAVVLATLVPLVYLGER
TH_0284|M.thermoresistible__bu --------------VLLAARPRRRTPILLLMPAAAVVTATLLPLVYLVQR
Mb2420c|M.bovis_AF2122/97 -----------------MTSLPAARYLVRSVALGYVFVLLIVPVALILWR
Rv2398c|M.tuberculosis_H37Rv -----------------MTSLPAARYLVRSVALGYVFVLLIVPVALILWR
MMAR_3716|M.marinum_M -----------------MTSSPLVRYLLRAIALTYVFALLVVPVGVILWR
MUL_3659|M.ulcerans_Agy99 -----------------MTSSPLVRYLLRAIALTYVFALLVVPVGVILWR
MAV_1784|M.avium_104 -----------------MTSSPGVRYGLRFVALAYIFVLLVIPVSLILWR
MAB_1654|M.abscessus_ATCC_1997 -----------------MTLSPAVKYALRILALLYVAVLLIVPLGLILWR
MLBr_02093|M.leprae_Br4923 MNLDTLDRPVKAVVLRPISVRRRIKNNVATMFFFTSFLIALVPLVWLLSV
::*: :
Mflv_3362|M.gilvum_PYR-GCK AFERGLPFVVDELLQPRTAA-------------MVGRSLLLVSVVTAACV
Mvan_3092|M.vanbaalenii_PYR-1 ALGRGWPFVVDELLQPRTAA-------------LVGRSLLLVAVVTAACV
MSMEG_3635|M.smegmatis_MC2_155 AAERGWAFVFDELFQPRTAA-------------LVGRSLALVALVTAACV
TH_0284|M.thermoresistible__bu ALERGWAFVLHELAQPRTAA-------------LIGRSLLLVVVVTAACV
Mb2420c|M.bovis_AF2122/97 TFEPGFGQFYAWISTPAAIS-------------ALNLSLLVVAIVVPLNV
Rv2398c|M.tuberculosis_H37Rv TFEPGFGQFYAWISTPAAIS-------------ALNLSLLVVAIVVPLNV
MMAR_3716|M.marinum_M TFEPGLGQFYDWISTPAAIS-------------ALHLSLLLVAIVVPLNV
MUL_3659|M.ulcerans_Agy99 TFEPGLGQFYDWISTPAAIS-------------ALHLSLLLVAIVVPLEV
MAV_1784|M.avium_104 TFRPGFGQFYAWVSTPAAIS-------------ALNLTLLVVAIVVPLNV
MAB_1654|M.abscessus_ATCC_1997 AFAPGLVQFWEYISTPAAIS-------------AFNLSLLVVAIVVPLNV
MLBr_02093|M.leprae_Br4923 VVARGFYAVTRFDWWTHSLRGVMPEEFAGGVYHALYGSLVQAVVAAIMAV
. * . . : . :* . :.. *
Mflv_3362|M.gilvum_PYR-GCK VLGVGLAVLVTRTDLR-GRRVLAVALTLPLAMP----SYLLAYLWVSA--
Mvan_3092|M.vanbaalenii_PYR-1 VLGVGLAVLVTRTNLR-GRRALAVSLTLPLAMP----SYLLAYLWVSA--
MSMEG_3635|M.smegmatis_MC2_155 VLGVGLAVLVSRTDLR-GRRVLAVVLTLPLAMP----SYLLAYLWVSA--
TH_0284|M.thermoresistible__bu VLGVGFAVLVNRTDLV-ARRVLAVALTLPLAIP----SYLLAFLWVSL--
Mb2420c|M.bovis_AF2122/97 IFGVTTALVLARNRFR-GKGVLQAIIDLPFAVSPVIVGVSLILLWGSAGA
Rv2398c|M.tuberculosis_H37Rv IFGVTTALVLARNRFR-GKGVLQAIIDLPFAVSPVIVGVSLILLWGSAGA
MMAR_3716|M.marinum_M IFGILTALLLARNRFR-GKGVLQAIIDLPFAVSPVIVGTALILLWGSAGA
MUL_3659|M.ulcerans_Agy99 IFGIPTALLLARNRFR-GKGVLQAIIDLPFAVSPVIVGTALILLWGSAGA
MAV_1784|M.avium_104 FFGIPTALVLARNRFR-GKGVLQAIIDLPFAVSPVIVGVALIVLWGSAGA
MAB_1654|M.abscessus_ATCC_1997 IFGVPTALVLARNKFR-GKGVLQAIIDLPFAVSPVIVGVALIVLWGSAGL
MLBr_02093|M.leprae_Br4923 PLGSMTAVYLVEYGSGPFVRVTTFMVDVLAGVPSIVAALFIFCLWIAT--
:* *: : . . : : .:. . : ** :
Mflv_3362|M.gilvum_PYR-GCK ---------FPTAAGFWGAALVLTLVSYPLVLLTTMAALARVDPAQEEVA
Mvan_3092|M.vanbaalenii_PYR-1 ---------YPTITGFWGAALVLTLVSYPLVLLTTMAALARVDPAQEEVA
MSMEG_3635|M.smegmatis_MC2_155 ---------YPQVAGFWGAALVLVLVSYPLVFLTTMAALSRVDPALEEVA
TH_0284|M.thermoresistible__bu ---------VPTISGFWGSTLVLVLVSYPLVMLTTMAALSRVDPAQEEVA
Mb2420c|M.bovis_AF2122/97 LGFVEQDLGFKIIFGLPGIVLASMFVTCPFVVREVEPVLHELGTDQEQAA
Rv2398c|M.tuberculosis_H37Rv LGFVEQDLGFKIIFGLPGIVLGSMFVTCPFVVREVEPVLHELGTDQEQAA
MMAR_3716|M.marinum_M LGFVERDLGIRIIFGLPGMVLASIFVTLPFVVREVEPVLHELGTDQEQAA
MUL_3659|M.ulcerans_Agy99 LDFVERDLGIRIIFGLPGMVLASIFVTLPFVVREVEPVLHELGTDQEQAA
MAV_1784|M.avium_104 LGFVEKDLGFKIIFGLPGIVLASIFVTLPFVVREVEPVLHELGTDQEEAA
MAB_1654|M.abscessus_ATCC_1997 LGFVENDWKFKIIFGLPGIVLASIFVTLPFVIREVEPVLHEIGDDQEQAA
MLBr_02093|M.leprae_Br4923 ---------LGFQQSAFAVSLALVLLMLPVVVRSTEEILRLVPDELREAC
. . * :: *.*. . * : .:..
Mflv_3362|M.gilvum_PYR-GCK RSLGLGGVATLFRVTLRQTRAAIAAGALLVALYVLSDFGAVAAMR-----
Mvan_3092|M.vanbaalenii_PYR-1 RSLGLGGVAVLFRVTLRQARPAIAAGALLVALYVLSDFGAVAAMR-----
MSMEG_3635|M.smegmatis_MC2_155 RSLGHGGFSVLFRVTLRQARGAITAGALLVALYVLSDFGAVAAMR-----
TH_0284|M.thermoresistible__bu RSLGLGGWRVLFRVTLRQSRAAITAGALLVALYVLSDFGAVAAMR-----
Mb2420c|M.bovis_AF2122/97 ATLGSGWWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAVIIVSSN---
Rv2398c|M.tuberculosis_H37Rv ATLGSGWWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAVIIVSSN---
MMAR_3716|M.marinum_M ATLGSGWWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAVLMVSSN---
MUL_3659|M.ulcerans_Agy99 ATLGSGWWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAVLMVSSN---
MAV_1784|M.avium_104 ATLGSGWWQTFWRITLPSIRWGLTYGIVLTIARSLGEYGAVLMVSSN---
MAB_1654|M.abscessus_ATCC_1997 ATLGANPWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAVIMVSSG---
MLBr_02093|M.leprae_Br4923 YALGIPKWKTIVRIVFPIATPGIISGVLLSIARVIGETAPVLVLVGYSRS
:** .: *:.: .: * :* :.: ..* :
Mflv_3362|M.gilvum_PYR-GCK ---------YEAFTWVIYGAYRSGFNPSRAAVLSLVLIVFAVLLVVGEHR
Mvan_3092|M.vanbaalenii_PYR-1 ---------FEAFTWVIYGAYRSGFNPSRAAVLSLVLMVFAVALVVAEHR
MSMEG_3635|M.smegmatis_MC2_155 ---------YEAFTWVIYGAYRSGFNPSRAAVLSLVLLVLAVLLVTAEAR
TH_0284|M.thermoresistible__bu ---------YEAFTWVIYGAYRSGFNPSRAAVLALVLLVFAVVLVLAEAR
Mb2420c|M.bovis_AF2122/97 -----LPGTSQTLTLLVSDRYHRGAEYGAYALSTLLMAVSVVVLIVQMVL
Rv2398c|M.tuberculosis_H37Rv -----LPGTSQTLTLLVSDRYHRGAEYGAYALSTLLMAVSVVVLIVQMVL
MMAR_3716|M.marinum_M -----LPGTSQTLTLLVSDRYHRGAEYGAYALSTLLMAVAVVVLIVQVAL
MUL_3659|M.ulcerans_Agy99 -----LPGTSQTLTLLVSDRYHRGAGYGAYALSTLLMAVAVVVLIVQVAL
MAV_1784|M.avium_104 -----LPGKSQTLTLLVSDRYNRGAEYGAYALSTLLMGVAVLVLVFQVVL
MAB_1654|M.abscessus_ATCC_1997 -----LPGKSQTLTLLVSDRHLRGDEYGAYALSVVLMAVALIVLVVQVVL
MLBr_02093|M.leprae_Br4923 INFDIFDGNMASLPLLIYTELTNPEYAGFLRVWGAALTLIILVAGINLFG
::. :: . : : : :
Mflv_3362|M.gilvum_PYR-GCK ARGVAAAGRIGGGAPRPAPVSRLGRWRWPALAVPAVVLTAALVVPGIELM
Mvan_3092|M.vanbaalenii_PYR-1 ARGRAAASRIGGGSPRPAPLNRLGPWSPLALLVPAAVLAAAVAVPTVELA
MSMEG_3635|M.smegmatis_MC2_155 ARGIAAA-RIGGGTPRPAPLNRLGRWWPAALVAPIAVLMAAIGLPTVILV
TH_0284|M.thermoresistible__bu ARGRAAASRVGGGTPRXX--------------------------------
Mb2420c|M.bovis_AF2122/97 DAHRARAVSEG---------------------------------------
Rv2398c|M.tuberculosis_H37Rv DARRARAVSEG---------------------------------------
MMAR_3716|M.marinum_M DVRRSRTTSRA---------------------------------------
MUL_3659|M.ulcerans_Agy99 DVRRSRTTSRA---------------------------------------
MAV_1784|M.avium_104 DARRGRAARQA---------------------------------------
MAB_1654|M.abscessus_ATCC_1997 DRNRAKTSK-----------------------------------------
MLBr_02093|M.leprae_Br4923 AAFRFLATRRCSGCR-----------------------------------
:
Mflv_3362|M.gilvum_PYR-GCK DWLLAAGMQWNVGEWLGALGSTVWLSAAAALVSTAAALPLGVLAARHRDR
Mvan_3092|M.vanbaalenii_PYR-1 GWLLTAGPRWDVAEWLDALGSTVWLSAAAAVVSTAAALPLGVLAARHRDR
MSMEG_3635|M.smegmatis_MC2_155 RWLLTAGARWDAQQWVDALTATLWLSFVTAIVCTFAALPLGVLAARYRDR
TH_0284|M.thermoresistible__bu ---XRSGPDWRR--------------------------------------
Mb2420c|M.bovis_AF2122/97 --------------------------------------------------
Rv2398c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3716|M.marinum_M --------------------------------------------------
MUL_3659|M.ulcerans_Agy99 --------------------------------------------------
MAV_1784|M.avium_104 --------------------------------------------------
MAB_1654|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02093|M.leprae_Br4923 --------------------------------------------------
Mflv_3362|M.gilvum_PYR-GCK VTRALEGASFLAHGLPSIVIAISMVSLGVLLLRPIYQREPLLILAYAVLF
Mvan_3092|M.vanbaalenii_PYR-1 ATRTLEGASYVAHGLPAIVIAISMVSLGVLLLRPIYQREPLLILAYTVLF
MSMEG_3635|M.smegmatis_MC2_155 VTRVLEGASYVAHGLPAIVIGISMVSLGVLVLRPLYQREPLLILAYAVLL
TH_0284|M.thermoresistible__bu --------------------------------------------------
Mb2420c|M.bovis_AF2122/97 --------------------------------------------------
Rv2398c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3716|M.marinum_M --------------------------------------------------
MUL_3659|M.ulcerans_Agy99 --------------------------------------------------
MAV_1784|M.avium_104 --------------------------------------------------
MAB_1654|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02093|M.leprae_Br4923 --------------------------------------------------
Mflv_3362|M.gilvum_PYR-GCK VPMAIGSIRASVQATPVRLEEVARSLGRPPVRAFATVTARVALPGIAAGA
Mvan_3092|M.vanbaalenii_PYR-1 VPMAIGSIRAAVEAAPIRLEEVARSLGRPPVRAFGAVTARVALPGIAAGA
MSMEG_3635|M.smegmatis_MC2_155 IPLAVGSVRAAIESTPARLEEVARSLGRNPLRAFTTVTARLSVPGIAAAA
TH_0284|M.thermoresistible__bu --------------------------------------------------
Mb2420c|M.bovis_AF2122/97 --------------------------------------------------
Rv2398c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3716|M.marinum_M --------------------------------------------------
MUL_3659|M.ulcerans_Agy99 --------------------------------------------------
MAV_1784|M.avium_104 --------------------------------------------------
MAB_1654|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02093|M.leprae_Br4923 --------------------------------------------------
Mflv_3362|M.gilvum_PYR-GCK ALVLLTCMKELPVTLLLHPTGTDTLATRLWGYSSISDYAAAAPYAAALLL
Mvan_3092|M.vanbaalenii_PYR-1 ALVLLTCMKELPVTLLLHPTGTDTLATRLWGYSSVSDYAAAAPYAAALLL
MSMEG_3635|M.smegmatis_MC2_155 ALVLLTCMKELPVTLLLHPTGTDTLATRLWGHSAVSDYAAAAPYAAALLV
TH_0284|M.thermoresistible__bu --------------------------------------------------
Mb2420c|M.bovis_AF2122/97 --------------------------------------------------
Rv2398c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3716|M.marinum_M --------------------------------------------------
MUL_3659|M.ulcerans_Agy99 --------------------------------------------------
MAV_1784|M.avium_104 --------------------------------------------------
MAB_1654|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02093|M.leprae_Br4923 --------------------------------------------------
Mflv_3362|M.gilvum_PYR-GCK FAAVPTAVLGLWSTRSAEVRVD
Mvan_3092|M.vanbaalenii_PYR-1 FAAVPTAVLGMWGTDDAGVRSD
MSMEG_3635|M.smegmatis_MC2_155 FAAIPTAVLGAWTAGRTGVRSD
TH_0284|M.thermoresistible__bu ----------------------
Mb2420c|M.bovis_AF2122/97 ----------------------
Rv2398c|M.tuberculosis_H37Rv ----------------------
MMAR_3716|M.marinum_M ----------------------
MUL_3659|M.ulcerans_Agy99 ----------------------
MAV_1784|M.avium_104 ----------------------
MAB_1654|M.abscessus_ATCC_1997 ----------------------
MLBr_02093|M.leprae_Br4923 ----------------------