For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADVSTAAATAVDAALLDALPPSEVPGRAAATRPGPVLTAGVVLLVVAAFVPIGYVAWSVVSLGPTRVWEL LARPRVLELFVNTVGLVVVTVPICIVLGIGAAWLVERSDIPGRAIWRPMFVAPLAVPAFINSYAWVSVVP TLHGFWAGILVASLSYFPFVYVPAAATLRRLDPAIEESARTLGSSPAAVFRRVVLPQLRLAVLGGGLLIG VHLLAEYGAFAMLRFSTFTTAIFEQFQATFDGAAGSTLAGVLMLLCLVLLVGEAGVRGNARYARLGSGAQ RTSLPHRLGSMTFPAVAALAALAALALGVPVWTILRWLWIGGSDVWALGEIVPSTVQTVALAAAAAALTT VLAFPFAWIAVRHRGFFARFVEGSNFVTSSMPGIVTALALVTVAIRFAPPLYQTATLVLAAYVLMFMPRA MVNLRSGLAQVPPTLEEASRSLGVSPTRTLLRVTLRLTAPAAAAGASLVFVAVATELTATLLLAPTGTNT LAMRFWSLSSELSYASAAPYALLLVVLAVPVTVVLFQQSSRAAAL*
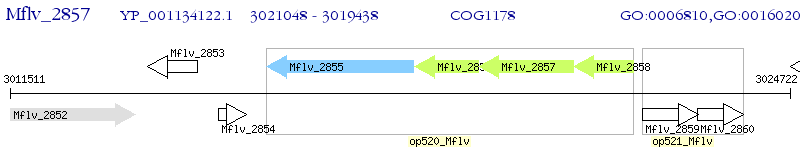
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2857 | - | - | 100% (536) | binding-protein-dependent transport systems inner membrane component |
| M. gilvum PYR-GCK | Mflv_3362 | - | 4e-94 | 38.45% (502) | binding-protein-dependent transport systems inner membrane |
| M. gilvum PYR-GCK | Mflv_2679 | - | 2e-14 | 23.21% (224) | sulfate ABC transporter, inner membrane subunit CysT |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2421c | cysT | 7e-14 | 24.69% (239) | sulfate-transport integral membrane protein ABC transporter |
| M. tuberculosis H37Rv | Rv2399c | cysT | 7e-14 | 24.69% (239) | sulfate-transport integral membrane protein ABC transporter |
| M. leprae Br4923 | MLBr_01088 | - | 2e-08 | 25.24% (206) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_1654 | - | 2e-16 | 26.14% (241) | sulfate ABC transporter, permease protein CysW |
| M. marinum M | MMAR_3716 | cysW | 2e-12 | 25.53% (235) | sulfate-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_1783 | cysT | 6e-15 | 26.92% (260) | sulfate ABC transporter, permease protein CysT |
| M. smegmatis MC2 155 | MSMEG_3635 | - | 3e-89 | 37.14% (490) | iron(III)-transport system permease protein SfuB |
| M. thermoresistible (build 8) | TH_0284 | - | 7e-43 | 37.82% (238) | iron(III)-transport system permease protein SfuB |
| M. ulcerans Agy99 | MUL_3659 | cysW | 8e-13 | 24.46% (233) | sulfate-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_3092 | - | 8e-93 | 38.05% (502) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3635|M.smegmatis_MC2_155 ------------------------------------MLVAAGAAVVLATL
Mvan_3092|M.vanbaalenii_PYR-1 ------------------------MPVITARSRPPALLLLPAALVAAGTL
TH_0284|M.thermoresistible__bu -----------------------VLLAARPRRRTPILLLMPAAAVVTATL
Mflv_2857|M.gilvum_PYR-GCK MADVSTAAATAVDAALLDALPPSEVPGRAAATRPGPVLTAGVVLLVVAAF
MMAR_3716|M.marinum_M --------------------------MTSSPLVRYLLRAIALTYVFALLV
MUL_3659|M.ulcerans_Agy99 --------------------------MTSSPLVRYLLRAIALTYVFALLV
MAB_1654|M.abscessus_ATCC_1997 --------------------------MTLSPAVKYALRILALLYVAVLLI
Mb2421c|M.bovis_AF2122/97 -----MTESLVGER----RAPQFRARLSGPAGPPSVRVGMAVVWLSVIVL
Rv2399c|M.tuberculosis_H37Rv -----MTESLVGER----RAPQFRARLSGPAGPPSVRVGMAVVWLSVIVL
MAV_1783|M.avium_104 --MTAITPDPLAIRPELGGDPGHQPVPGRGRGTTALRVGVATVWLSVIVL
MLBr_01088|M.leprae_Br4923 ----------------MGIEKVKVTAGVRVGARRATIWVIIDTFVVGYAL
: .
MSMEG_3635|M.smegmatis_MC2_155 VPLVYLGERAAERG--------------WAFVFDELFQPRTAALVGRSLA
Mvan_3092|M.vanbaalenii_PYR-1 IPLVYLVERALGRG--------------WPFVVDELLQPRTAALVGRSLL
TH_0284|M.thermoresistible__bu LPLVYLVQRALERG--------------WAFVLHELAQPRTAALIGRSLL
Mflv_2857|M.gilvum_PYR-GCK VPIGYVAWSVVSLG--------------PTRVWELLARPRVLELFVNTVG
MMAR_3716|M.marinum_M VPVGVILWRTFEPG--------------LGQFYDWISTPAAISALHLSLL
MUL_3659|M.ulcerans_Agy99 VPVGVILWRTFEPG--------------LGQFYDWISTPAAISALHLSLL
MAB_1654|M.abscessus_ATCC_1997 VPLGLILWRAFAPG--------------LVQFWEYISTPAAISAFNLSLL
Mb2421c|M.bovis_AF2122/97 LPLAAIVWQAAGGG--------------WRAFWLAVSSHAAMESFRVTLT
Rv2399c|M.tuberculosis_H37Rv LPLAAIVWQAAGGG--------------WRAFWLAVSSHAAMESFRVTLT
MAV_1783|M.avium_104 LPLAAIAWQSAGGG--------------WHVFWLAVTSNAALESFRVTLT
MLBr_01088|M.leprae_Br4923 LPVLWILSLSLKPASTVKDGKLIPSLVTFDNYRGIFRSDLFSSALINSIG
:*: : . . . ::
MSMEG_3635|M.smegmatis_MC2_155 LVALVTAACVVLGVGLAVLVSRTDLRGRRVLAVVLTLPLAMPSYLLAYLW
Mvan_3092|M.vanbaalenii_PYR-1 LVAVVTAACVVLGVGLAVLVTRTNLRGRRALAVSLTLPLAMPSYLLAYLW
TH_0284|M.thermoresistible__bu LVVVVTAACVVLGVGFAVLVNRTDLVARRVLAVALTLPLAIPSYLLAFLW
Mflv_2857|M.gilvum_PYR-GCK LVVVTVPICIVLGIGAAWLVERSDIPGRAIWRPMFVAPLAVPAFINSYAW
MMAR_3716|M.marinum_M LVAIVVPLNVIFGILTALLLARNRFRGKGVLQAIIDLPFAVSPVIVGTAL
MUL_3659|M.ulcerans_Agy99 LVAIVVPLEVIFGIPTALLLARNRFRGKGVLQAIIDLPFAVSPVIVGTAL
MAB_1654|M.abscessus_ATCC_1997 VVAIVVPLNVIFGVPTALVLARNKFRGKGVLQAIIDLPFAVSPVIVGVAL
Mb2421c|M.bovis_AF2122/97 ISTAVTVINLVFGLLIAWVLVRDDFAGKRIVDAIIDLPFALPTIVASLVM
Rv2399c|M.tuberculosis_H37Rv ISTAVTVINLVFGLLIAWVLVRDDFAGKRIVDAIIDLPFALPTIVASLVM
MAV_1783|M.avium_104 ISAGVTVLNLVFGLVIAWVLVRDEFPGKRLVDAIIDLPFALPTIVASLVM
MLBr_01088|M.leprae_Br4923 IGLTTTVIAVMFGAMAAYAIARLAFPGKRLLVGTTLLVTMFPAISLVTPL
: .. :::* * : * : .: ...
MSMEG_3635|M.smegmatis_MC2_155 VSAYPQV---------------AGFWGAALVLVLVSYPLVFLTTMAALSR
Mvan_3092|M.vanbaalenii_PYR-1 VSAYPTI---------------TGFWGAALVLTLVSYPLVLLTTMAALAR
TH_0284|M.thermoresistible__bu VSLVPTI---------------SGFWGSTLVLVLVSYPLVMLTTMAALSR
Mflv_2857|M.gilvum_PYR-GCK VSVVPTL---------------HGFWAGILVASLSYFPFVYVPAAATLRR
MMAR_3716|M.marinum_M ILLWGSAGALGFVERDLGIRIIFGLPGMVLASIFVTLPFVVREVEPVLHE
MUL_3659|M.ulcerans_Agy99 ILLWGSAGALDFVERDLGIRIIFGLPGMVLASIFVTLPFVVREVEPVLHE
MAB_1654|M.abscessus_ATCC_1997 IVLWGSAGLLGFVENDWKFKIIFGLPGIVLASIFVTLPFVIREVEPVLHE
Mb2421c|M.bovis_AF2122/97 LALYGNN-------SPVGLHFQHTATGVGVALAFVTLPFVVRAVQPVLLE
Rv2399c|M.tuberculosis_H37Rv LALYGNN-------SPVGLHFQHTATGVGVALAFVTLPFVVRAVQPVLLE
MAV_1783|M.avium_104 LALYGNN-------SPVGLHLQHTAAGVAVALAFVTLPFVVRAVQPVLLE
MLBr_01088|M.leprae_Br4923 FNIERRVG------------LFDTWAGLILPYITFALPLAIYTLSAFFAE
. . : *:. . : .
MSMEG_3635|M.smegmatis_MC2_155 VDPALEEVARSLGHGGFSVLFRVTLRQARGAITAGALLVALYVLSDFGAV
Mvan_3092|M.vanbaalenii_PYR-1 VDPAQEEVARSLGLGGVAVLFRVTLRQARPAIAAGALLVALYVLSDFGAV
TH_0284|M.thermoresistible__bu VDPAQEEVARSLGLGGWRVLFRVTLRQSRAAITAGALLVALYVLSDFGAV
Mflv_2857|M.gilvum_PYR-GCK LDPAIEESARTLGSSPAAVFRRVVLPQLRLAVLGGGLLIGVHLLAEYGAF
MMAR_3716|M.marinum_M LGTDQEQAAATLGSGWWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAV
MUL_3659|M.ulcerans_Agy99 LGTDQEQAAATLGSGWWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAV
MAB_1654|M.abscessus_ATCC_1997 IGDDQEQAAATLGANPWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAV
Mb2421c|M.bovis_AF2122/97 IDRETEEAAASLGANGAKIFTSVVLPSLTPALLSGAGLAFSRAIGEFGSV
Rv2399c|M.tuberculosis_H37Rv IDRETEEAAASLGANGAKIFTSVVLPSLTPALLSGAGLAFSRAIGEFGSV
MAV_1783|M.avium_104 LDRETEEAAASLGASGPKIFTSVVLPALLPALLSGAGLAFSRAIGEFGSV
MLBr_01088|M.leprae_Br4923 IPWDLEKAAKMDGATSGQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLA
: *: * * : : : .: . * :
MSMEG_3635|M.smegmatis_MC2_155 AAMR------YEAFTWVIYGAYRSGFNPSRAAVLSLVLLVLAVLLVTAEA
Mvan_3092|M.vanbaalenii_PYR-1 AAMR------FEAFTWVIYGAYRSGFNPSRAAVLSLVLMVFAVALVVAEH
TH_0284|M.thermoresistible__bu AAMR------YEAFTWVIYGAYRSGFNPSRAAVLALVLLVFAVVLVLAEA
Mflv_2857|M.gilvum_PYR-GCK AMLR------FSTFTTAIFEQFQATFDGAAGSTLAGVLMLLCLVLLVGEA
MMAR_3716|M.marinum_M LMVSSNLPGTSQTLTLLVSDRYHRGAEYGAYALSTLLMAVAVVVLIVQVA
MUL_3659|M.ulcerans_Agy99 LMVSSNLPGTSQTLTLLVSDRYHRGAGYGAYALSTLLMAVAVVVLIVQVA
MAB_1654|M.abscessus_ATCC_1997 IMVSSGLPGKSQTLTLLVSDRHLRGDEYGAYALSVVLMAVALIVLVVQVV
Mb2421c|M.bovis_AF2122/97 VLIGGAVPGKTEVSSQWIRTLIENDDRTGAAAISVVLLSISFIVLLILRV
Rv2399c|M.tuberculosis_H37Rv VLIGGAVPGKTEVSSQWIRTLIENDDRTGAAAISVVLLSISFIVLLILRV
MAV_1783|M.avium_104 VLIGGAVPGKTEVSSQWIRTLIENDDRTGAAAISIVLLAISFVVLLILRV
MLBr_01088|M.leprae_Br4923 LSLTSTKAAITAPVAITNFTGS-SQFEEPTGSIAAGAIVITAPIIVFVWI
: : : : : ::
MSMEG_3635|M.smegmatis_MC2_155 RARG-IAAARIGGGTPRPAPLNRLGRWWPAALVAPIAVLMAAIGLPTVIL
Mvan_3092|M.vanbaalenii_PYR-1 RARGRAAASRIGGGSPRPAPLNRLGPWSPLALLVPAAVLAAAVAVPTVEL
TH_0284|M.thermoresistible__bu RARGRAAASRVGGGTPRXX-------------------------------
Mflv_2857|M.gilvum_PYR-GCK GVRGNARYARLGSGAQRTSLPHRLGSMTFPAVAALAALAALALGVPVWTI
MMAR_3716|M.marinum_M LDVRRSRTTSRA--------------------------------------
MUL_3659|M.ulcerans_Agy99 LDVRRSRTTSRA--------------------------------------
MAB_1654|M.abscessus_ATCC_1997 LDRNRAKTSK----------------------------------------
Mb2421c|M.bovis_AF2122/97 VGARAAKREEMAA-------------------------------------
Rv2399c|M.tuberculosis_H37Rv VGARAAKREEMAA-------------------------------------
MAV_1783|M.avium_104 VGGRAAKREELAA-------------------------------------
MLBr_01088|M.leprae_Br4923 FQRRIVAGLTSGAVKG----------------------------------
MSMEG_3635|M.smegmatis_MC2_155 VRWLLTAGA-RWDAQQWVDALTATLWLSFVTAIVCTFAALPLGVLAARYR
Mvan_3092|M.vanbaalenii_PYR-1 AGWLLTAGP-RWDVAEWLDALGSTVWLSAAAAVVSTAAALPLGVLAARHR
TH_0284|M.thermoresistible__bu ----XRSGP-DWRR------------------------------------
Mflv_2857|M.gilvum_PYR-GCK LRWLWIGGSDVWALGEIVPSTVQTVALAAAAAALTTVLAFPFAWIAVRHR
MMAR_3716|M.marinum_M --------------------------------------------------
MUL_3659|M.ulcerans_Agy99 --------------------------------------------------
MAB_1654|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2421c|M.bovis_AF2122/97 --------------------------------------------------
Rv2399c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1783|M.avium_104 --------------------------------------------------
MLBr_01088|M.leprae_Br4923 --------------------------------------------------
MSMEG_3635|M.smegmatis_MC2_155 DRVTRVLEGASYVAHGLPAIVIGISMVSLGVLVLRPLYQREPLLILAYAV
Mvan_3092|M.vanbaalenii_PYR-1 DRATRTLEGASYVAHGLPAIVIAISMVSLGVLLLRPIYQREPLLILAYTV
TH_0284|M.thermoresistible__bu --------------------------------------------------
Mflv_2857|M.gilvum_PYR-GCK GFFARFVEGSNFVTSSMPGIVTALALVTVAIRFAPPLYQTATLVLAAYVL
MMAR_3716|M.marinum_M --------------------------------------------------
MUL_3659|M.ulcerans_Agy99 --------------------------------------------------
MAB_1654|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2421c|M.bovis_AF2122/97 --------------------------------------------------
Rv2399c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1783|M.avium_104 --------------------------------------------------
MLBr_01088|M.leprae_Br4923 --------------------------------------------------
MSMEG_3635|M.smegmatis_MC2_155 LLIPLAVGSVRAAIESTPARLEEVARSLGRNPLRAFTTVTARLSVPGIAA
Mvan_3092|M.vanbaalenii_PYR-1 LFVPMAIGSIRAAVEAAPIRLEEVARSLGRPPVRAFGAVTARVALPGIAA
TH_0284|M.thermoresistible__bu --------------------------------------------------
Mflv_2857|M.gilvum_PYR-GCK MFMPRAMVNLRSGLAQVPPTLEEASRSLGVSPTRTLLRVTLRLTAPAAAA
MMAR_3716|M.marinum_M --------------------------------------------------
MUL_3659|M.ulcerans_Agy99 --------------------------------------------------
MAB_1654|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2421c|M.bovis_AF2122/97 --------------------------------------------------
Rv2399c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1783|M.avium_104 --------------------------------------------------
MLBr_01088|M.leprae_Br4923 --------------------------------------------------
MSMEG_3635|M.smegmatis_MC2_155 AAALVLLTCMKELPVTLLLHPTGTDTLATRLWGHSAVSDYAAAAPYAAAL
Mvan_3092|M.vanbaalenii_PYR-1 GAALVLLTCMKELPVTLLLHPTGTDTLATRLWGYSSVSDYAAAAPYAAAL
TH_0284|M.thermoresistible__bu --------------------------------------------------
Mflv_2857|M.gilvum_PYR-GCK GASLVFVAVATELTATLLLAPTGTNTLAMRFWSLSSELSYASAAPYALLL
MMAR_3716|M.marinum_M --------------------------------------------------
MUL_3659|M.ulcerans_Agy99 --------------------------------------------------
MAB_1654|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2421c|M.bovis_AF2122/97 --------------------------------------------------
Rv2399c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1783|M.avium_104 --------------------------------------------------
MLBr_01088|M.leprae_Br4923 --------------------------------------------------
MSMEG_3635|M.smegmatis_MC2_155 LVFAAIPTAVLGAWTAGRTGVRSD
Mvan_3092|M.vanbaalenii_PYR-1 LLFAAVPTAVLGMWGTDDAGVRSD
TH_0284|M.thermoresistible__bu ------------------------
Mflv_2857|M.gilvum_PYR-GCK VVLAVPVTVVLFQQSSRAAAL---
MMAR_3716|M.marinum_M ------------------------
MUL_3659|M.ulcerans_Agy99 ------------------------
MAB_1654|M.abscessus_ATCC_1997 ------------------------
Mb2421c|M.bovis_AF2122/97 ------------------------
Rv2399c|M.tuberculosis_H37Rv ------------------------
MAV_1783|M.avium_104 ------------------------
MLBr_01088|M.leprae_Br4923 ------------------------