For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YPPSTAGSLRLGIDGGQMKVLITGGTGFVGAWTAKAAQDAGHQVRFLVRNPDRLTTSAAEIGADISDHVI GDIADGEATAAALDGCDAVIHCAAMVSTDPSRADEMLHTNLEGARNILGGAAHAGIDPIVHVSSFTALFR PDLDRLHADLPVVGGSDGYGRSKAAVEAYARGLQDGGAPVNITYPGMVLGPPAGDQFGEAADGVEASVKM RGVPGRGAGWIVIDVRDLADLHVALLEPGRGPRRYMAGGQRVSVSELASMIGNAADQSILVYPVPDVALR SAGRLLDVVGPYLPFETPINSAAMQYYTQMPESDDTPGARDFGLTQRDPAVTIADTFAGLRAVGRL*
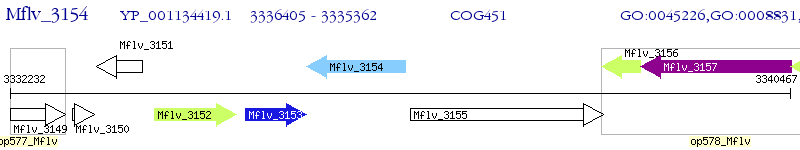
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3154 | - | - | 100% (347) | NAD-dependent epimerase/dehydratase |
| M. gilvum PYR-GCK | Mflv_1564 | - | 3e-53 | 36.17% (329) | NAD-dependent epimerase/dehydratase |
| M. gilvum PYR-GCK | Mflv_2375 | - | 3e-11 | 26.63% (338) | NAD-dependent epimerase/dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3752 | - | 1e-126 | 63.91% (327) | putative oxidoreductase |
| M. tuberculosis H37Rv | Rv3725 | - | 4e-97 | 65.99% (247) | oxidoreductase |
| M. leprae Br4923 | MLBr_01942 | - | 2e-09 | 27.53% (178) | putative cholesterol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2971c | - | 1e-122 | 64.00% (325) | putative oxidoreductase |
| M. marinum M | MMAR_5252 | - | 1e-126 | 64.92% (325) | oxidoreductase |
| M. avium 104 | MAV_1849 | - | 1e-13 | 26.20% (313) | dihydroflavonol-4-reductase family protein |
| M. smegmatis MC2 155 | MSMEG_5909 | - | 4e-53 | 37.65% (324) | oxidoreductase |
| M. thermoresistible (build 8) | TH_3827 | - | 3e-12 | 31.07% (177) | PUTATIVE putative dehydrogenase |
| M. ulcerans Agy99 | MUL_4328 | - | 1e-126 | 64.92% (325) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3375 | - | 1e-170 | 89.36% (329) | NAD-dependent epimerase/dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3154|M.gilvum_PYR-GCK MYPPSTAGSLRLGIDGGQMKVLITGGTGFVGAWTAKAAQDAGHQVRFLVR
Mvan_3375|M.vanbaalenii_PYR-1 ------------------MKVLVTGGTGFVGAWTAKAAQQAGHQVRFLVR
Mb3752|M.bovis_AF2122/97 -------------MQNATMRVLVTGGTGFVGGWTAKAIADAGHSVRFLVR
Rv3725|M.tuberculosis_H37Rv -------------MQNATMRVLVTGGTGFVGGWTAKAIADAGHSVRFLVR
MMAR_5252|M.marinum_M -------------MHNRCMHVLVTGGTGFVGGWTAKAIADAGHSVRFLVR
MUL_4328|M.ulcerans_Agy99 -------------MHNRCMHVLVTGGTGFVGGWTAKAIADAGHSVRFLVR
MAB_2971c|M.abscessus_ATCC_199 ------------------MRVLVTGGTGFVGGWTAKAMAEAGHKVRFLVR
MSMEG_5909|M.smegmatis_MC2_155 -------------MSEESVRIAVTGGTGYVGAHTVRGLLTAGHEVRLLVA
MAV_1849|M.avium_104 -----------------MAVKLVIGASGFLGSHVTRQLVERGERVRVLLR
TH_3827|M.thermoresistible__bu ----MTDEDRIPCGAMSRPSVFITGANGFIGRSLGAHFAENGYDVR----
MLBr_01942|M.leprae_Br4923 ----MLRGVGNASLTTELGRVLVTGGSGFVGANLVTALLERGYQVR----
: *..*::* * **
Mflv_3154|M.gilvum_PYR-GCK NPDRLTTSAAEIG-ADISDHVIGDIADGEATAAALDGCDAVIHCAAMVST
Mvan_3375|M.vanbaalenii_PYR-1 NPDRLTTSAEKIG-VDISDHAVGDIADAAATAAALDGCDAVIHCAAMVST
Mb3752|M.bovis_AF2122/97 NPARLKTSVAKLG-VDVSDFAVADISDRDSVREALNGCDAVVHSAALVAT
Rv3725|M.tuberculosis_H37Rv NPARLKTSVAKLG-VDVSDFAVADISDRDSVREALNGCDAVVHSAALVAT
MMAR_5252|M.marinum_M NPAKLESSVAKLG-VDVSDFIIGDIKDRDLVREALTGCDAVVHSAALVAT
MUL_4328|M.ulcerans_Agy99 NPAKLESSVAKLG-VDVSDFIIGDIKDRDLVREALTGCDAVVHSAALVAT
MAB_2971c|M.abscessus_ATCC_199 KEARLHTTVATLG-VDVSDYAVGDITDRASVEAALDGCDAVVHSAAMVST
MSMEG_5909|M.smegmatis_MC2_155 PGCEGEPVIDKLAEAGEVETLVGDIRDSGTVDRLIKGCDSVIHAAGVVGT
MAV_1849|M.avium_104 RTSS-TVALDDLD----IERRYGDVFDDAVLRDALDGCDDVYYCVVDTRA
TH_3827|M.thermoresistible__bu ---GVDLAADPAR-----NVVAGDVVNPRPWKSHLQGCAVVLHTAAVVSN
MLBr_01942|M.leprae_Br4923 ---SFDRAPMPLPQHPQLEVLQGDITDATVCTTAMDSIDTVFHTAAIIEL
: .*: : : . * : .
Mflv_3154|M.gilvum_PYR-GCK D------PSRADEMLHTNLEGARNILGGAAHAGIDPIVHVSSFTALFRPD
Mvan_3375|M.vanbaalenii_PYR-1 D------PSRADEMLHTNLEGARNVLGEAARVGIDPIVHVSSFTALFRPD
Mb3752|M.bovis_AF2122/97 D------PRETSRMLSTNMAGAQNVLGQAVELGMDPIVHVSSFTALFRPN
Rv3725|M.tuberculosis_H37Rv D------PRETSRMLSTNMAGAQNVLGQAVELGMDPIVHVSSFTALFRPN
MMAR_5252|M.marinum_M D------QRQTQDMLSTNMEGARNVLGQAVALGLDPIVHVSSFTALFHPG
MUL_4328|M.ulcerans_Agy99 D------QRQTQDMLSTNMEGARNVLGQAVALGLDPIVHVSSFTALFHPG
MAB_2971c|M.abscessus_ATCC_199 D------PAEADRMMAINLEGTRNVLGAAVDRGLDPIVHVSSITALFRPG
MSMEG_5909|M.smegmatis_MC2_155 D------KSRTQLMWEINAHATEAVLIRAAEAGLDPIVSVSSYSSLFPPP
MAV_1849|M.avium_104 W------LRDPAPLFRTNVEGLRQVLDTAVGADLRRFVFTSTIGTIALSE
TH_3827|M.thermoresistible__bu -------AVAAGQQWQANVLGTRRVLEAAARAGVERVVHFSSVRAYGDLG
MLBr_01942|M.leprae_Br4923 MGGASVTDEYRQRSYTVNVGGTENLLRAGQKSGVKRFVYTASNSVVMGGT
* . . :* . .: .* ::
Mflv_3154|M.gilvum_PYR-GCK LDR-LHADLPVVG-GSDGYGRSKAAVEAYARGLQDG----GAPVNITYPG
Mvan_3375|M.vanbaalenii_PYR-1 LDV-LHADLPVVG-GSDGYGRSKAVVEAYARGLQDG----GAPVNITYPG
Mb3752|M.bovis_AF2122/97 LAT-LSADLPVAG-GTDGYGQSKAQIEIYARGLQDA----GAPVNITYPG
Rv3725|M.tuberculosis_H37Rv LAT-LSADLPVAG-GTDGYGQSKAQIEIYARGLQDA----GAPVNITYPG
MMAR_5252|M.marinum_M LQT-LSADLPVVG-GSDGYGTSKAQVEIYARGLQDA----GAPVNITYPG
MUL_4328|M.ulcerans_Agy99 LQT-LSADLPVVG-GSDGYGTSKAQVEIYARGLQDA----GAPVNITYPG
MAB_2971c|M.abscessus_ATCC_199 LET-FAADLPAAG-GSDGYGQSKARVELYVRALQDS----GAPVTITYPG
MSMEG_5909|M.smegmatis_MC2_155 DGV-ISADTPPVA-GRSPYAQTKAYADRAARRLQDT----GAPVVVTYPS
MAV_1849|M.avium_104 DGLPVSEDEPFNW-ADRGGGYIRSRVEAENLVLQYARE-RGLPAVAMCVS
TH_3827|M.thermoresistible__bu FPDGVKETWPLRA-DGRAYVDTKVAAEATAFAAHASG---EIAVTVVRPG
MLBr_01942|M.leprae_Br4923 PITGGDETMPYTKRFNDLYTETKVVAEKFVLSQNGVPDGETMLTCSIRPS
* : : : . .
Mflv_3154|M.gilvum_PYR-GCK MVLGPPAGDQFGEAADGVEASVKMRGVPGRGAGWIVIDVRDLADLHVALL
Mvan_3375|M.vanbaalenii_PYR-1 MVLGPPAGDQFGEAAEGVEASVKMRGVPGRSAAWIVIDVRDLADLHVALL
Mb3752|M.bovis_AF2122/97 MVLGPPVGDQFGEAGEGVRSALWMHVIPGRGAAWLIVDVRDVAALHAALL
Rv3725|M.tuberculosis_H37Rv MVLGPPVGDQFGEAGEGVRSALWMHVIPGRGAAWLIVDVRDVAALHAALL
MMAR_5252|M.marinum_M MVLGPPVGNQFGEAGEGVKAALEIRTIPGRNAAWLIIDVRDVAAVHAALL
MUL_4328|M.ulcerans_Agy99 MVLGPPVGNQFGEAGEGVKAALEIRTIPGRNAAWLIIDVRDVAAVHAALL
MAB_2971c|M.abscessus_ATCC_199 MVLGPPVGDQFGEAAEGVRWVLLTRSVPGRSAAWLVVDVRDLAALHTALL
MSMEG_5909|M.smegmatis_MC2_155 SVVGPAWFTAPGVTERGWGPIVKARVAPRLRGGMQMIDVRDVADVHVALM
MAV_1849|M.avium_104 NTYGPGDFQPTPHGSLVAAAGKGKMPVYVKDMSMEVVGIEDAARALILAA
TH_3827|M.thermoresistible__bu DVYGPGSRPWTIIPVEEIKKGRLILPAGGQGVFSPVYIDDLIAGILLAAT
MLBr_01942|M.leprae_Br4923 GIWGRGDQTMFRKAFESVVSGHVKVLIGSKNAKLDNSYVHNLVHGLILAA
* .
Mflv_3154|M.gilvum_PYR-GCK E---PG--RGPRRYMAGGQR-VSVSELASMIGNAADQSILVYPVPDVALR
Mvan_3375|M.vanbaalenii_PYR-1 E---PG--RGPRRYMAGGQR-VPVDMLASMIGTAAGRSLAVYPVPDVALR
Mb3752|M.bovis_AF2122/97 E---SG--RGPRRYTAGGHR-IPVPELAKILGEVAGTTMLAVPVPDSALR
Rv3725|M.tuberculosis_H37Rv E---SG--RGPRRYTAGGHR-IPVPELAKILGGSPAPRCWPSRCPIPRCV
MMAR_5252|M.marinum_M E---PG--RGPRRYMVGGHR-IPVPELAKMLGQVAETPIVSVPIPDSVLR
MUL_4328|M.ulcerans_Agy99 E---PG--RGPRRYMVGGHR-IPVPELAKMLGQVAETPIVSVPIPDSVLR
MAB_2971c|M.abscessus_ATCC_199 E---PS--RGPRRYTAGGHR-LEVGELVDLLTEAAGSAVLAVPVPDSLLR
MSMEG_5909|M.smegmatis_MC2_155 K---PG--RGPHRYVCGGVL-VSFNEWIDILEQGVGRPIRRIPLSPAMFH
MAV_1849|M.avium_104 E---KG--RVGERYIVSERY-ISARELYTTAAEAGGARPPRIGIPLKVMY
TH_3827|M.thermoresistible__bu H---PD--AAGQDFNLPGVEPVTCKTFFGHYTRMLNKPPPRT-VPTRVAI
MLBr_01942|M.leprae_Br4923 EHLVPGGTAPGQAYFINDGEPINFFDFMGPIIKACGENWPRVRISGRLVR
. . . : : .
Mflv_3154|M.gilvum_PYR-GCK -SAGRLLDVVGPYLPFETPINSAAMQYYTQMPESDDTPGARDFGLTQRD-
Mvan_3375|M.vanbaalenii_PYR-1 -SAGRLLDVVGPFLPFETPINSAAMQYYTQMPESDDTPSRRDLGITQRD-
Mb3752|M.bovis_AF2122/97 -VAGSVLDQAGPYLPFNTPFTAAGMQYYTQMPEPDDSPSEKELGITYRD-
Rv3725|M.tuberculosis_H37Rv -SRDRCWIKPGP-------------ICLSILRSPRQVCS-----TTHRC-
MMAR_5252|M.marinum_M -VAGSLLDLAGPYLPFDNPFTAAGMQYYTQMPDSDDSPSELELGVTYRD-
MUL_4328|M.ulcerans_Agy99 -VAGSLLDLAGPYLPFDNPFTAAGMQYYTQMPDSDDSPSELELGVTYRD-
MAB_2971c|M.abscessus_ATCC_199 -AAGTVMDQIGRYVPWETPMTEAGMQYYTQMPASDDTPSERELGITYRD-
MSMEG_5909|M.smegmatis_MC2_155 -AIGWVSDLLGNVLPLGDGISYEAAMLLTAATPTDDAKTLADLGIPAWRS
MAV_1849|M.avium_104 -ALGLCGDVAAKVLRRDMLLSTLSVRLMHIMSPMDHSKAERELGWRPEP-
TH_3827|M.thermoresistible__bu -ALATAAGTVERFRGRSSELNAETVYYLTRAGGYSRAKAERVLGWVPQVS
MLBr_01942|M.leprae_Br4923 NVMAVWQRLHFGFGLPKPPMEPLAVERVYLDNYFSIEKAHKELGYRPLFT
Mflv_3154|M.gilvum_PYR-GCK PAVTIADTFAGLRAVGRL-------------------
Mvan_3375|M.vanbaalenii_PYR-1 PAETIADTVEGLRRVGRL-------------------
Mb3752|M.bovis_AF2122/97 PRDTVADTVTALRGLGS--------------------
Rv3725|M.tuberculosis_H37Rv RSPTIRRAKKN--------------------------
MMAR_5252|M.marinum_M PRTTLADTVAALTA-----------------------
MUL_4328|M.ulcerans_Agy99 PRTTLADTVAALTA-----------------------
MAB_2971c|M.abscessus_ATCC_199 PRETLADTVVSLRAGRTTSKLWGLWPFSE--------
MSMEG_5909|M.smegmatis_MC2_155 PRAAVLESFGR--------------------------
MAV_1849|M.avium_104 IHDAIRRAVAFYQTR----------------------
TH_3827|M.thermoresistible__bu LDEGMARTEAWLRTEGYLD------------------
MLBr_01942|M.leprae_Br4923 TEQAMAECLPYYTELFEQIKVAAKPHLASIAATPRPE
: