For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTLTHVDGAPLPSFTPGSHIVVECEAPGRPTNAYSLTGDGIAPDAYVISVLRRADEITSGPSGSQWIHDE LAVGDTLIARPPRSAFAPVLRARRHLLVAAGIGITPMVSHLRSAKLWGRETRLYYIHRPGRAAYVDTIRE LTDHASIHTERAQFATDVSVALADQPMGTHLYVCGPAGFIEFVSATAAALGWPASRIHIEHFGSEALDPG EPFSVRVRSTGEEFPVEPGVSLLEALERRGFDIPNLCRRGVCGECRIPVSGSVITHRDLYLTEDDKVAGD SLMACVSRADGSQTLEVAL
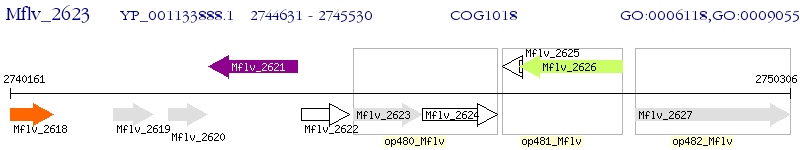
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2623 | - | - | 100% (299) | ferredoxin |
| M. gilvum PYR-GCK | Mflv_3426 | - | 3e-32 | 33.33% (300) | ferredoxin |
| M. gilvum PYR-GCK | Mflv_1462 | - | 2e-18 | 28.90% (301) | ferredoxin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2798c | - | 7e-23 | 30.85% (295) | oxidoreductase |
| M. tuberculosis H37Rv | Rv2776c | - | 7e-23 | 30.85% (295) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2381 | - | 9e-38 | 33.56% (292) | oxidoreductase |
| M. marinum M | MMAR_1933 | - | 6e-26 | 31.35% (303) | oxidoreductase |
| M. avium 104 | MAV_3669 | - | 4e-27 | 32.33% (300) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_4638 | - | 3e-97 | 58.53% (299) | vanillate O-demethylase oxidoreductase |
| M. thermoresistible (build 8) | TH_3804 | - | 5e-40 | 35.08% (305) | PUTATIVE ferredoxin |
| M. ulcerans Agy99 | MUL_2159 | - | 2e-27 | 31.68% (303) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3958 | - | 1e-135 | 78.41% (301) | ferredoxin |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2798c|M.bovis_AF2122/97 --------------------------------------------------
Rv2776c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1933|M.marinum_M ------MAVSESSWTSRPADLYGRSHRDRFATALWGVRTMFGGFARFSRW
MUL_2159|M.ulcerans_Agy99 ------MAVSESSWTSRPADLYGRSHRDRFATALWGVRTMFGGFARFSRW
MAV_3669|M.avium_104 MSIRRAASRAPSIWASRPADLYGRRDHDRFTAVLWGVRAVFEGFASVSRW
TH_3804|M.thermoresistible__bu -------MRMLARLRATPPSSSGRIRHDLLLGVADATISSLWKVSEVLR-
MAB_2381|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_2623|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3958|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4638|M.smegmatis_MC2_155 --------------------------------------------------
Mb2798c|M.bovis_AF2122/97 -------MRRTNPAVVTKRELVAP--DVVALTLADPGGGLLPAWSPGGHI
Rv2776c|M.tuberculosis_H37Rv -------MRRTNPAVVTKRELVAP--DVVALTLADPGGGLLPAWSPGGHI
MMAR_1933|M.marinum_M NPSRVTPVGRTHRARVVERELVAP--DVVALTLADPDGGLLPSWSPGGHI
MUL_2159|M.ulcerans_Agy99 NPSRVTPVGRTHRARVVERELVAP--DVVALTLADPDGGLLPSWSPGGHI
MAV_3669|M.avium_104 HPARVKPVQRKLTAVVTKREFAAP--DVVALTLADPDGGLLPSWTPGAHI
TH_3804|M.thermoresistible__bu TPRLPAVDAEPITLTVRDRRVVAHDQDVIALTLVAADGGQLPPWRPGAHI
MAB_2381|M.abscessus_ATCC_1997 ----MNDTGATLKVAVRQLRWEAD--GVVSLQLSSACNEPLPAWEPGAHV
Mflv_2623|M.gilvum_PYR-GCK ------------------------------MTLTHVDGAPLPSFTPGSHI
Mvan_3958|M.vanbaalenii_PYR-1 --------MTTLKLTVAAIDDTIA--GIRTLTLTHVDEAPLPSFTPGSHL
MSMEG_4638|M.smegmatis_MC2_155 ------MTQPVMQLVITEIDDSVP--GVRTLRLAHPEGRALPSYPPGSHV
: * **.: **.*:
Mb2798c|M.bovis_AF2122/97 DVQLPS--GRRRQYSLCGVPGRRTDYRIAIRRIADGGG----GSIEMHEA
Rv2776c|M.tuberculosis_H37Rv DVQLPS--GRRRQYSLCGVPGRRTDYRIAIRRIADGGG----GSIEMHEA
MMAR_1933|M.marinum_M DVQLPS--GRRRQYSLCGPPGRRTDYRIAVRRIADGGG----GSIEMHAA
MUL_2159|M.ulcerans_Agy99 DVQLPS--RRRRQYSLCGPPGRRTDYRIAVRRIADGGG----GSIEMHAA
MAV_3669|M.avium_104 DVLLPS--GRRRQYSLCGPPGHRGEYRIAVRRIADGGG----GSIEMHEA
TH_3804|M.thermoresistible__bu DVTLPS--GRVRQYSLCGDPAVRDRYRIAVRRIPDGGG----GSVEVHDA
MAB_2381|M.abscessus_ATCC_1997 DLVLPT--GIERQYSLCGPVDDRSVYQVGVRRERTSRG----GSEYVHAF
Mflv_2623|M.gilvum_PYR-GCK VVECEAPGRPTNAYSLTGDGIAPDAYVISVLRRADEITSGPSGSQWIHDE
Mvan_3958|M.vanbaalenii_PYR-1 VVECGARNGPANAYSLTGDGIAPAAYVISVLR-AGTSGEAPSGSRWIHDE
MSMEG_4638|M.smegmatis_MC2_155 VLQCGD---VANAYSLTGDGVSPMRYVVSVLLCPNGSG----GSRWIHDE
: . *** * * :.: ** :*
Mb2798c|M.bovis_AF2122/97 FDVGDTCEFEGPRNAFHL-----GLAERDVLFVIGGIGVTPILPMIRAAE
Rv2776c|M.tuberculosis_H37Rv FDVGDTCEFEGPRNAFHL-----GLAERDVLFVIGGIGVTPILPMIRAAE
MMAR_1933|M.marinum_M YQPGDILVFEGPRNAFYL-----GAAEHDMLFVIGGIGVTPILPMVALAH
MUL_2159|M.ulcerans_Agy99 YQPGDILVFEGPRNAFYL-----GAAEHDMLFVIGGIGVTPILPMVALAH
MAV_3669|M.avium_104 FGVGDTVEFEGPRNAFYL-----GTAEHDVLFVIGGIGVTPILPMIRVAA
TH_3804|M.thermoresistible__bu LHLGARVTTHGPRNAFPLSVPGYGSPARRLRFIAGGIGITPILPMLAEAQ
MAB_2381|M.abscessus_ATCC_1997 LRPGQQLRIKGPRNNFGL------HRADGYLFIAAGIGITPILPMIRQAQ
Mflv_2623|M.gilvum_PYR-GCK LAVGDTLIARPPRSAFAP-----VLRARRHLLVAAGIGITPMVSHLRSAK
Mvan_3958|M.vanbaalenii_PYR-1 LAVGDTVLARPPRSAFAP-----ILRARRHLLVAAGIGITPMVSHLRSAR
MSMEG_4638|M.smegmatis_MC2_155 VRIGETIDVSAPRSAFAP-----VHKARRHLLVAAGIGITPMMSHLYSAK
* **. * :: .***:**::. : *
Mb2798c|M.bovis_AF2122/97 QRGIDWRAIYAGRGREYMPFLDEVVAVAPGRVTVWADDEHGRFASVDELL
Rv2776c|M.tuberculosis_H37Rv QRGIDWRAIYAGRGREYMPFLDEVVAVAPGRVTVWADDEHGRFASVDELL
MMAR_1933|M.marinum_M QRGVNWRAMYAGRSREYMPLLDELVAMAPERVSVWADDEHGRFAQVEDLV
MUL_2159|M.ulcerans_Agy99 QRGVNWRAVYAGRSREYMPLLDELVAMAPEGVSVWADDEHGRFAQVEDLV
MAV_3669|M.avium_104 GRGIDWRAIYTGRSREYMPLLDEVVSVDPQRVTVWADDEHGRFPTAAELL
TH_3804|M.thermoresistible__bu AFGVPWSMIYAGRSRDSLPFLEELTRYG-DRVRVRTDDVAG-LPTAAELL
MAB_2381|M.abscessus_ATCC_1997 DWGADWTLYYGGHTAASMAFLEELRQYG-PRVHLYPADEVGRIPLR-ELT
Mflv_2623|M.gilvum_PYR-GCK LWGRETRLYYIHRPGR-AAYVDTIRELT--DHASIHTERAQFATDVSVAL
Mvan_3958|M.vanbaalenii_PYR-1 LWGRDARLLYIHRPGR-GAYVDTVRSLT--DHASIHTLREEFVAELSTAL
MSMEG_4638|M.smegmatis_MC2_155 RWGADARLLYIHRAGR-GAYLDEVEQLG--MPARVCTDRDRFHSELTAAL
* * : . :: : .
Mb2798c|M.bovis_AF2122/97 AGAGPTTAVYVCGPPGMLEAVRVARNQH--ADAPLHYERFSPPPVVDG--
Rv2776c|M.tuberculosis_H37Rv AGAGPTTAVYVCGPPGMLEAVRVARNQH--ADAPLHYERFSPPPVVDG--
MMAR_1933|M.marinum_M AGAGPKTAVYVCGPPPMLEAVRAARAEY--PCAPLHYERFSPPPVVDG--
MUL_2159|M.ulcerans_Agy99 AGAGLKTAVYVCGPPPMLEAVRAARAEY--PCAPLHYERFSPPPVVDG--
MAV_3669|M.avium_104 AAAGPATAVYVCGPSAMLEAVWIARNEH--AGAPLHYERFSPAPVVDG--
TH_3804|M.thermoresistible__bu GACPDGTAVYACGPPAMLTAIRAELAGR--RNVELHFERFAAPPVVDG--
MAB_2381|M.abscessus_ATCC_1997 TDVRPGLQIYACGPTELISDLTAAVSHW--PADTLHVERFKPIPCAPAED
Mflv_2623|M.gilvum_PYR-GCK ADQPMGTHLYVCGPAGFIEFVSATAAALGWPASRIHIEHFGSEALDPG--
Mvan_3958|M.vanbaalenii_PYR-1 ADQPLGTHLYVCGPASFIDFVAATATELGWPASRIHFEHFGAGALDPG--
MSMEG_4638|M.smegmatis_MC2_155 ISQPIGTHLYVCGPGTFMDDVIAQARELGWPPSRIHSERFGVDALDAG--
:*.*** :: : :* *:* . .
Mb2798c|M.bovis_AF2122/97 VPFELELARSRRVLRVPANRSALDVMLDWDPTTAYSCQQGFCGTCKVRVL
Rv2776c|M.tuberculosis_H37Rv VPFELELARSRRVLRVPANRSALDVMLDWDPTTAYSCQQGFCGTCKVRVL
MMAR_1933|M.marinum_M VPFELELARTGEVLSIPANRTALDVMLDRDPTTAYSCQQGFCGTCKVKVL
MUL_2159|M.ulcerans_Agy99 VPFELELARTGEVLSIPANRTALDVMLDRDPTTAYSCQQGFCGTCKVKVL
MAV_3669|M.avium_104 VPFELELARSRQVLAVPANRTALDVMLDRDATTPYSCRQGFCGTCRVKVL
TH_3804|M.thermoresistible__bu RPFSVRIASTGAEVPVGADETLLAALGRAGVQAPYSCQQGFCGTCRTRVL
MAB_2381|M.abscessus_ATCC_1997 KPVDVTCAASRQTIAVPPGQSILAAVEQAGIQVSASCRAGVCGSCETAVL
Mflv_2623|M.gilvum_PYR-GCK EPFSVRVRSTGEEFPVEPGVSLLEALERRGFDIPNLCRRGVCGECRIPVS
Mvan_3958|M.vanbaalenii_PYR-1 EPFTVRVPSTADEFTVEAGVSLLEALESRGFAIPNLCRRGVCGECRVPVS
MSMEG_4638|M.smegmatis_MC2_155 EPFEVELAGTGQTLTVPSGVSLLEALEKNGQNVPNLCRQGVCGECRIPVA
*. : : . : .. : * .: . . *: *.** *. *
Mb2798c|M.bovis_AF2122/97 AGQVDRRGRIIE-----GDNEMLVCVSRAVS-GRVVIDA
Rv2776c|M.tuberculosis_H37Rv AGQVDRRGRIIE-----GDNEMLVCVSRAVS-GRVVIDA
MMAR_1933|M.marinum_M AGQVDHRGRAAQ-----GDDEMLVCVSRGHS-ARVVIDA
MUL_2159|M.ulcerans_Agy99 AGQVDHRGRAAQ-----GDDEMLVCVSRGHS-ARVVIDA
MAV_3669|M.avium_104 AGQVDHRGRTDP-----GQDGMLVCVSRADG-GRLVIDA
TH_3804|M.thermoresistible__bu DGTVEHRDRLLTEPE-RAAGLMLTCVSRAPAGERLTLDL
MAB_2381|M.abscessus_ATCC_1997 EGVPDHRDDILSEEDRAAGDRMYICVSRALT-PRLVLDL
Mflv_2623|M.gilvum_PYR-GCK GSVITHRDLYLTEDDKVAGDSLMACVSRADGSQTLEVAL
Mvan_3958|M.vanbaalenii_PYR-1 GGAITHRDLYLSDDEKRAGNSMMACVSRA-ASQSLELKL
MSMEG_4638|M.smegmatis_MC2_155 RGDILHRDTYLSDDEKQAGDALMACVSRA-AGERLELAL
. :*. . . : ****. : :