For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGVLGRLRYDVTIGLADAALHGMFATSAAMRRIRPPDGVPETFPLTVTSRRVVARDQDVVELTLSGDDLP RWHPGAHLDLHLPSGRIRQYSLCGEPTAGHYRVAVRRIPDGGGGSVEVHDALPVGSTVRTHGPRNAFPLA VPGYGSPARRFRFIAGGIGITPILPMLALARSLGVDWSMVYTGRSADALPFVDEVAAFGSPVEIRTDDRR GLPTAAELLGDCPADTSVYACGPAPMLTAVRDALVGRDDVELHFERFAAPPVVDGRPFRVAAAGVTVDVG VDETLLAALGRAGVTTPYSCRQGFCGTCRTRVLSGEVDHRDTLLTDAERAAGLMLPCVSRAAEGTVLALD L
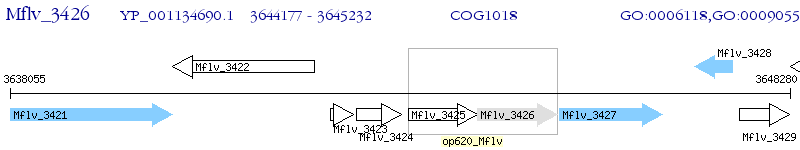
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3426 | - | - | 100% (351) | ferredoxin |
| M. gilvum PYR-GCK | Mflv_2623 | - | 3e-32 | 33.33% (300) | ferredoxin |
| M. gilvum PYR-GCK | Mflv_2468 | - | 1e-18 | 28.79% (323) | ferredoxin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2798c | - | 9e-70 | 47.32% (317) | oxidoreductase |
| M. tuberculosis H37Rv | Rv2776c | - | 9e-70 | 47.32% (317) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4020c | - | 1e-102 | 55.46% (339) | oxidoreductase |
| M. marinum M | MMAR_1933 | - | 5e-67 | 44.78% (335) | oxidoreductase |
| M. avium 104 | MAV_3669 | - | 4e-72 | 47.93% (338) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_3676 | - | 1e-135 | 65.73% (356) | phenoxybenzoate dioxygenase beta subunit |
| M. thermoresistible (build 8) | TH_3804 | - | 1e-135 | 67.42% (353) | PUTATIVE ferredoxin |
| M. ulcerans Agy99 | MUL_2159 | - | 1e-66 | 44.78% (335) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3180 | - | 1e-160 | 78.80% (349) | ferredoxin |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3426|M.gilvum_PYR-GCK ------------------MGVLGRLRYDV---TIGLADAALHGMFATSAA
Mvan_3180|M.vanbaalenii_PYR-1 ------MTRLFSQFRQTPPSPSGRLRHDV---MLGIAETAIAGMFAASAA
MSMEG_3676|M.smegmatis_MC2_155 ---------MLSQYRQLPPSTNGRFRHDP---MLAMAQIAIKTMWGVTRH
TH_3804|M.thermoresistible__bu -------MRMLARLRATPPSSSGRIRHDL---LLGVADATISSLWKVSEV
MAB_4020c|M.abscessus_ATCC_199 ---MTHALPMATQTGRHPLAWGMRVLDVVGPTAVAALGKTLPAVLRLSPL
Mb2798c|M.bovis_AF2122/97 --------------------------------------------------
Rv2776c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1933|M.marinum_M ------MAVSESSWTSRPADLYGRSHRDRFATALWGVRTMFGGFARFSRW
MUL_2159|M.ulcerans_Agy99 ------MAVSESSWTSRPADLYGRSHRDRFATALWGVRTMFGGFARFSRW
MAV_3669|M.avium_104 MSIRRAASRAPSIWASRPADLYGRRDHDRFTAVLWGVRAVFEGFASVSRW
Mflv_3426|M.gilvum_PYR-GCK MRRIRPPDGVPETFPLTVTSRRVVARDQDVVELTLSGDD---LPRWHPGA
Mvan_3180|M.vanbaalenii_PYR-1 VRRVRMPAAGPDTIPLTVTGRRVVAHDQDVVELTLAGDR---LPRWFPGA
MSMEG_3676|M.smegmatis_MC2_155 IRRPSPPPALDRTIGLRVVDRRVVAHDQDVIALTLAADDGGALPRWYPGA
TH_3804|M.thermoresistible__bu LRTPRLPAVDAEPITLTVRDRRVVAHDQDVIALTLVAADGGQLPPWRPGA
MAB_4020c|M.abscessus_ATCC_199 LGPVRPPKWVDRSLHLTVRERSVVAADQDVVALSLGAHDGGELPRWRPGA
Mb2798c|M.bovis_AF2122/97 ---------MRRTNPAVVTKRELVAP--DVVALTLADPGGGLLPAWSPGG
Rv2776c|M.tuberculosis_H37Rv ---------MRRTNPAVVTKRELVAP--DVVALTLADPGGGLLPAWSPGG
MMAR_1933|M.marinum_M NPSRVTP--VGRTHRARVVERELVAP--DVVALTLADPDGGLLPSWSPGG
MUL_2159|M.ulcerans_Agy99 NPSRVTP--VGRTHRARVVERELVAP--DVVALTLADPDGGLLPSWSPGG
MAV_3669|M.avium_104 HPARVKP--VQRKLTAVVTKREFAAP--DVVALTLADPDGGLLPSWTPGA
* * ..* **: *:* ** * **.
Mflv_3426|M.gilvum_PYR-GCK HLDLHLPSGRIRQYSLCGEPTAG-HYRVAVRRIPDGGGGSVEVHDALPVG
Mvan_3180|M.vanbaalenii_PYR-1 HLDVHLPSGRVRQYSLCGDPDSA-EYRIAVRRIPDGGGGSVEVHDALEIG
MSMEG_3676|M.smegmatis_MC2_155 HIDLHLPSGRIRQYSLCGDPAAG-TYRIAVRRIPEGGGGSVEIHDTVHIG
TH_3804|M.thermoresistible__bu HIDVTLPSGRVRQYSLCGDPAVRDRYRIAVRRIPDGGGGSVEVHDALHLG
MAB_4020c|M.abscessus_ATCC_199 HLDVTLPSGAVRQYSLCGDPRDRFSYRIAVRRIPNGNGGSIELHDGVSVG
Mb2798c|M.bovis_AF2122/97 HIDVQLPSGRRRQYSLCGVPGRRTDYRIAIRRIADGGGGSIEMHEAFDVG
Rv2776c|M.tuberculosis_H37Rv HIDVQLPSGRRRQYSLCGVPGRRTDYRIAIRRIADGGGGSIEMHEAFDVG
MMAR_1933|M.marinum_M HIDVQLPSGRRRQYSLCGPPGRRTDYRIAVRRIADGGGGSIEMHAAYQPG
MUL_2159|M.ulcerans_Agy99 HIDVQLPSRRRRQYSLCGPPGRRTDYRIAVRRIADGGGGSIEMHAAYQPG
MAV_3669|M.avium_104 HIDVLLPSGRRRQYSLCGPPGHRGEYRIAVRRIADGGGGSIEMHEAFGVG
*:*: *** ******* * **:*:***.:*.***:*:* *
Mflv_3426|M.gilvum_PYR-GCK STVRTHGPRNAFPLAVPGYG-SPARRFRFIAGGIGITPILPMLALARSLG
Mvan_3180|M.vanbaalenii_PYR-1 STVHTHGPRNAFPLTVPGYG-SPARRFRFIAGGIGITPILPMLGLARRLG
MSMEG_3676|M.smegmatis_MC2_155 SRLTTHGPRNAFPLTVPGYG-SPTQRFRFIAGGIGITPILPMLDLAQRLG
TH_3804|M.thermoresistible__bu ARVTTHGPRNAFPLSVPGYG-SPARRLRFIAGGIGITPILPMLAEAQAFG
MAB_4020c|M.abscessus_ATCC_199 DVLGVKGPRNAFPLATTGHLNTGTRRFHFVAGGIGITPILPMLAVATELG
Mb2798c|M.bovis_AF2122/97 DTCEFEGPRNAFHLGLAERD------VLFVIGGIGVTPILPMIRAAEQRG
Rv2776c|M.tuberculosis_H37Rv DTCEFEGPRNAFHLGLAERD------VLFVIGGIGVTPILPMIRAAEQRG
MMAR_1933|M.marinum_M DILVFEGPRNAFYLGAAEHD------MLFVIGGIGVTPILPMVALAHQRG
MUL_2159|M.ulcerans_Agy99 DILVFEGPRNAFYLGAAEHD------MLFVIGGIGVTPILPMVALAHQRG
MAV_3669|M.avium_104 DTVEFEGPRNAFYLGTAEHD------VLFVIGGIGVTPILPMIRVAAGRG
.****** * . . *: ****:******: * *
Mflv_3426|M.gilvum_PYR-GCK VDWSMVYTGRSADALPFVDEVAAFG-SPVEIRTDDRRG-LPTAAELLGDC
Mvan_3180|M.vanbaalenii_PYR-1 VDWSMVYTGRSVDSLPFVDEVTAFG-AAVDIRTDDRHG-LPAAAELLGDC
MSMEG_3676|M.smegmatis_MC2_155 VDWSMIYAGRHPDSLPFLSELQRYG-DRIGIRTDDRDG-LPGASDLLGEC
TH_3804|M.thermoresistible__bu VPWSMIYAGRSRDSLPFLEELTRYG-DRVRVRTDDVAG-LPTAAELLGAC
MAB_4020c|M.abscessus_ATCC_199 LPWTMTYTGRSRESIPFRDELARYG-DRITIRTDDEDG-LPTPADLLPPL
Mb2798c|M.bovis_AF2122/97 IDWRAIYAGRGREYMPFLDEVVAVAPGRVTVWADDEHGRFASVDELLAGA
Rv2776c|M.tuberculosis_H37Rv IDWRAIYAGRGREYMPFLDEVVAVAPGRVTVWADDEHGRFASVDELLAGA
MMAR_1933|M.marinum_M VNWRAMYAGRSREYMPLLDELVAMAPERVSVWADDEHGRFAQVEDLVAGA
MUL_2159|M.ulcerans_Agy99 VNWRAVYAGRSREYMPLLDELVAMAPEGVSVWADDEHGRFAQVEDLVAGA
MAV_3669|M.avium_104 IDWRAIYTGRSREYMPLLDEVVSVDPQRVTVWADDEHGRFPTAAELLAAA
: * *:** : :*: .*: : : :** * :. :*:
Mflv_3426|M.gilvum_PYR-GCK PADTSVYACGPAPMLTAVRDALVGRDDVELHFERFAAPPVVDGRPFR--V
Mvan_3180|M.vanbaalenii_PYR-1 ADGTAVYACGPAPMLTAVRAALVGRDDVELHFERFAAPPVTDGRPFQATV
MSMEG_3676|M.smegmatis_MC2_155 PGGTTVYACGPAPMLTMLRTALVGRDDVELHFERFAAPPVVDGTEFSVRI
TH_3804|M.thermoresistible__bu PDGTAVYACGPPAMLTAIRAELAGRRNVELHFERFAAPPVVDGRPFSVRI
MAB_4020c|M.abscessus_ATCC_199 HPDLAVYCCGPAPMLNVVRDAVAECDGAELHFERFSAPPIEHGVPFQVQL
Mb2798c|M.bovis_AF2122/97 GPTTAVYVCGPPGMLEAVRVARNQHADAPLHYERFSPPPVVDGVPFELEL
Rv2776c|M.tuberculosis_H37Rv GPTTAVYVCGPPGMLEAVRVARNQHADAPLHYERFSPPPVVDGVPFELEL
MMAR_1933|M.marinum_M GPKTAVYVCGPPPMLEAVRAARAEYPCAPLHYERFSPPPVVDGVPFELEL
MUL_2159|M.ulcerans_Agy99 GLKTAVYVCGPPPMLEAVRAARAEYPCAPLHYERFSPPPVVDGVPFELEL
MAV_3669|M.avium_104 GPATAVYVCGPSAMLEAVWIARNEHAGAPLHYERFSPAPVVDGVPFELEL
:** ***. ** : . **:***:..*: .* * :
Mflv_3426|M.gilvum_PYR-GCK AAAGVTVDVGVDETLLAALGRAGVTTPYSCRQGFCGTCRTRVL----SGE
Mvan_3180|M.vanbaalenii_PYR-1 ASTGQQVDVGADETLLAALRRAGVDASYSCQQGFCGTCRTRVL----AGS
MSMEG_3676|M.smegmatis_MC2_155 ASTGQTVPVAADETLLAALNRAGVSAPYSCQQGFCGTCRVRALPRDGDGA
TH_3804|M.thermoresistible__bu ASTGAEVPVGADETLLAALGRAGVQAPYSCQQGFCGTCRTRVL----DGT
MAB_4020c|M.abscessus_ATCC_199 GIGGPVLEVGADQSALAAVLTSRPGTPHSCRQGFCGTCKVKVL----AGR
Mb2798c|M.bovis_AF2122/97 ARSRRVLRVPANRSALDVMLDWDPTTAYSCQQGFCGTCKVRVL----AGQ
Rv2776c|M.tuberculosis_H37Rv ARSRRVLRVPANRSALDVMLDWDPTTAYSCQQGFCGTCKVRVL----AGQ
MMAR_1933|M.marinum_M ARTGEVLSIPANRTALDVMLDRDPTTAYSCQQGFCGTCKVKVL----AGQ
MUL_2159|M.ulcerans_Agy99 ARTGEVLSIPANRTALDVMLDRDPTTAYSCQQGFCGTCKVKVL----AGQ
MAV_3669|M.avium_104 ARSRQVLAVPANRTALDVMLDRDATTPYSCRQGFCGTCRVKVL----AGQ
. : : .:.: * .: :.:**:*******:.:.* *
Mflv_3426|M.gilvum_PYR-GCK VDHRDTLLTDAERAAGLMLPCVSRAAEGTVLALDL
Mvan_3180|M.vanbaalenii_PYR-1 VEHRDTLLTGPERAAGLMLTCVSRAGDGSGLTLDL
MSMEG_3676|M.smegmatis_MC2_155 VQHRDTLLTEPERAAGYLLTCVSRAGNGQRLTIEL
TH_3804|M.thermoresistible__bu VEHRDRLLTEPERAAGLMLTCVSRAPAGERLTLDL
MAB_4020c|M.abscessus_ATCC_199 PDHRDSLLTETQRAEGQMLLCVSRAD-GERLVLDI
Mb2798c|M.bovis_AF2122/97 VDRRGRIIEGDN----EMLVCVSRAV-SGRVVIDA
Rv2776c|M.tuberculosis_H37Rv VDRRGRIIEGDN----EMLVCVSRAV-SGRVVIDA
MMAR_1933|M.marinum_M VDHRGRAAQGDD----EMLVCVSRGH-SARVVIDA
MUL_2159|M.ulcerans_Agy99 VDHRGRAAQGDD----EMLVCVSRGH-SARVVIDA
MAV_3669|M.avium_104 VDHRGRTDPGQD----GMLVCVSRAD-GGRLVIDA
::*. : :* ****. . :.::