For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAVSESSWTSRPADLYGRSHRDRFATALWGVRTMFGGFARFSRWNPSRVTPVGRTHRARVVERELVAPDV VALTLADPDGGLLPSWSPGGHIDVQLPSGRRRQYSLCGPPGRRTDYRIAVRRIADGGGGSIEMHAAYQPG DILVFEGPRNAFYLGAAEHDMLFVIGGIGVTPILPMVALAHQRGVNWRAMYAGRSREYMPLLDELVAMAP ERVSVWADDEHGRFAQVEDLVAGAGPKTAVYVCGPPPMLEAVRAARAEYPCAPLHYERFSPPPVVDGVPF ELELARTGEVLSIPANRTALDVMLDRDPTTAYSCQQGFCGTCKVKVLAGQVDHRGRAAQGDDEMLVCVSR GHSARVVIDA
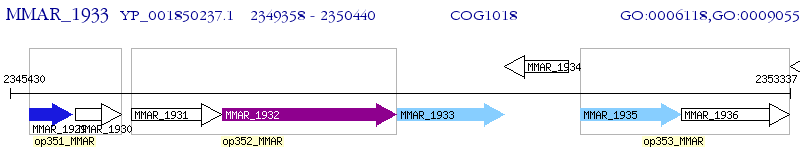
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1933 | - | - | 100% (360) | oxidoreductase |
| M. marinum M | MMAR_5066 | hmp | 3e-18 | 27.73% (321) | flavodoxin reductase Hmp |
| M. marinum M | MMAR_5043 | fdxB | 2e-15 | 25.36% (351) | electron transfer protein FdxB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2798c | - | 1e-145 | 79.94% (309) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_3426 | - | 4e-67 | 44.78% (335) | ferredoxin |
| M. tuberculosis H37Rv | Rv2776c | - | 1e-145 | 79.94% (309) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4020c | - | 4e-80 | 50.94% (320) | oxidoreductase |
| M. avium 104 | MAV_4119 | - | 1e-157 | 74.17% (360) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_3676 | - | 2e-72 | 43.67% (371) | phenoxybenzoate dioxygenase beta subunit |
| M. thermoresistible (build 8) | TH_3804 | - | 1e-73 | 49.36% (312) | PUTATIVE ferredoxin |
| M. ulcerans Agy99 | MUL_2159 | - | 0.0 | 98.89% (360) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3180 | - | 9e-72 | 48.40% (312) | ferredoxin |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3676|M.smegmatis_MC2_155 ------MLSQYRQLPPSTNGRFRHDP---MLAMAQIAIKTMWGVTRHIRR
TH_3804|M.thermoresistible__bu ----MRMLARLRATPPSSSGRIRHDL---LLGVADATISSLWKVSEVLRT
Mflv_3426|M.gilvum_PYR-GCK ---------------MGVLGRLRYDV---TIGLADAALHGMFATSAAMRR
Mvan_3180|M.vanbaalenii_PYR-1 ---MTRLFSQFRQTPPSPSGRLRHDV---MLGIAETAIAGMFAASAAVRR
MAB_4020c|M.abscessus_ATCC_199 MTHALPMATQTGRHPLAWGMRVLDVVGPTAVAALGKTLPAVLRLSPLLGP
MMAR_1933|M.marinum_M ---MAVSESSWTSRPADLYGRSHRDRFATALWGVRTMFGGFARFSRWNPS
MUL_2159|M.ulcerans_Agy99 ---MAVSESSWTSRPADLYGRSHRDRFATALWGVRTMFGGFARFSRWNPS
Mb2798c|M.bovis_AF2122/97 --------------------------------------------------
Rv2776c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4119|M.avium_104 ---MTMAQAIWESIPADLYGRRKRDRMYTALYGVGALFGGLASASRWTPA
MSMEG_3676|M.smegmatis_MC2_155 PSPPPALDRTIGLRVVDRRVVAHDQDVIALTLAADDGGALPRWYPGAHID
TH_3804|M.thermoresistible__bu PRLPAVDAEPITLTVRDRRVVAHDQDVIALTLVAADGGQLPPWRPGAHID
Mflv_3426|M.gilvum_PYR-GCK IRPPDGVPETFPLTVTSRRVVARDQDVVELTLSGDD---LPRWHPGAHLD
Mvan_3180|M.vanbaalenii_PYR-1 VRMPAAGPDTIPLTVTGRRVVAHDQDVVELTLAGDR---LPRWFPGAHLD
MAB_4020c|M.abscessus_ATCC_199 VRPPKWVDRSLHLTVRERSVVAADQDVVALSLGAHDGGELPRWRPGAHLD
MMAR_1933|M.marinum_M RVTP--VGRTHRARVVERELVAP--DVVALTLADPDGGLLPSWSPGGHID
MUL_2159|M.ulcerans_Agy99 RVTP--VGRTHRARVVERELVAP--DVVALTLADPDGGLLPSWSPGGHID
Mb2798c|M.bovis_AF2122/97 ------MRRTNPAVVTKRELVAP--DVVALTLADPGGGLLPAWSPGGHID
Rv2776c|M.tuberculosis_H37Rv ------MRRTNPAVVTKRELVAP--DVVALTLADPGGGLLPAWSPGGHID
MAV_4119|M.avium_104 RVVP--VRRTTTAVVTQREELAP--DVVAFTLADPHGGLLPSWTPGAHID
. * * :* **: ::* ** * **.*:*
MSMEG_3676|M.smegmatis_MC2_155 LHLPSGRIRQYSLCGDPAAG-TYRIAVRRIPEGGGGSVEIHDTVHIGSRL
TH_3804|M.thermoresistible__bu VTLPSGRVRQYSLCGDPAVRDRYRIAVRRIPDGGGGSVEVHDALHLGARV
Mflv_3426|M.gilvum_PYR-GCK LHLPSGRIRQYSLCGEPTAG-HYRVAVRRIPDGGGGSVEVHDALPVGSTV
Mvan_3180|M.vanbaalenii_PYR-1 VHLPSGRVRQYSLCGDPDSA-EYRIAVRRIPDGGGGSVEVHDALEIGSTV
MAB_4020c|M.abscessus_ATCC_199 VTLPSGAVRQYSLCGDPRDRFSYRIAVRRIPNGNGGSIELHDGVSVGDVL
MMAR_1933|M.marinum_M VQLPSGRRRQYSLCGPPGRRTDYRIAVRRIADGGGGSIEMHAAYQPGDIL
MUL_2159|M.ulcerans_Agy99 VQLPSRRRRQYSLCGPPGRRTDYRIAVRRIADGGGGSIEMHAAYQPGDIL
Mb2798c|M.bovis_AF2122/97 VQLPSGRRRQYSLCGVPGRRTDYRIAIRRIADGGGGSIEMHEAFDVGDTC
Rv2776c|M.tuberculosis_H37Rv VQLPSGRRRQYSLCGVPGRRTDYRIAIRRIADGGGGSIEMHEAFDVGDTC
MAV_4119|M.avium_104 VRLPSGRRRQYSLCGPPGRRTDYRIAVRRIEDGGGGSIEMHDTFRVGDAL
: *** ******* * **:*:*** :*.***:*:* *
MSMEG_3676|M.smegmatis_MC2_155 TTHGPRNAFPLTVPGYG-SPTQRFRFIAGGIGITPILPMLDLAQRLGVDW
TH_3804|M.thermoresistible__bu TTHGPRNAFPLSVPGYG-SPARRLRFIAGGIGITPILPMLAEAQAFGVPW
Mflv_3426|M.gilvum_PYR-GCK RTHGPRNAFPLAVPGYG-SPARRFRFIAGGIGITPILPMLALARSLGVDW
Mvan_3180|M.vanbaalenii_PYR-1 HTHGPRNAFPLTVPGYG-SPARRFRFIAGGIGITPILPMLGLARRLGVDW
MAB_4020c|M.abscessus_ATCC_199 GVKGPRNAFPLATTGHLNTGTRRFHFVAGGIGITPILPMLAVATELGLPW
MMAR_1933|M.marinum_M VFEGPRNAFYLGAAEHD------MLFVIGGIGVTPILPMVALAHQRGVNW
MUL_2159|M.ulcerans_Agy99 VFEGPRNAFYLGAAEHD------MLFVIGGIGVTPILPMVALAHQRGVNW
Mb2798c|M.bovis_AF2122/97 EFEGPRNAFHLGLAERD------VLFVIGGIGVTPILPMIRAAEQRGIDW
Rv2776c|M.tuberculosis_H37Rv EFEGPRNAFHLGLAERD------VLFVIGGIGVTPILPMIRAAEQRGIDW
MAV_4119|M.avium_104 EFEGPRNAFYLVTDERD------VLFVIGGIGVTPILPMIQTAQQHGINW
.****** * . *: ****:******: * *: *
MSMEG_3676|M.smegmatis_MC2_155 SMIYAGRHPDSLPFLSELQRYG-DRIGIRTDDRDG-LPGASDLLGECPGG
TH_3804|M.thermoresistible__bu SMIYAGRSRDSLPFLEELTRYG-DRVRVRTDDVAG-LPTAAELLGACPDG
Mflv_3426|M.gilvum_PYR-GCK SMVYTGRSADALPFVDEVAAFG-SPVEIRTDDRRG-LPTAAELLGDCPAD
Mvan_3180|M.vanbaalenii_PYR-1 SMVYTGRSVDSLPFVDEVTAFG-AAVDIRTDDRHG-LPAAAELLGDCADG
MAB_4020c|M.abscessus_ATCC_199 TMTYTGRSRESIPFRDELARYG-DRITIRTDDEDG-LPTPADLLPPLHPD
MMAR_1933|M.marinum_M RAMYAGRSREYMPLLDELVAMAPERVSVWADDEHGRFAQVEDLVAGAGPK
MUL_2159|M.ulcerans_Agy99 RAVYAGRSREYMPLLDELVAMAPEGVSVWADDEHGRFAQVEDLVAGAGLK
Mb2798c|M.bovis_AF2122/97 RAIYAGRGREYMPFLDEVVAVAPGRVTVWADDEHGRFASVDELLAGAGPT
Rv2776c|M.tuberculosis_H37Rv RAIYAGRGREYMPFLDEVVAVAPGRVTVWADDEHGRFASVDELLAGAGPT
MAV_4119|M.avium_104 RAVYAGRSRDYMPLLDEVVAVAPERVSVWADDERGRIPTAADLLAEAGPA
*:** : :*: .*: . : : :** * :. :*:
MSMEG_3676|M.smegmatis_MC2_155 TTVYACGPAPMLTMLRTALVGRDDVELHFERFAAPPVVDGTEFSVRIAST
TH_3804|M.thermoresistible__bu TAVYACGPPAMLTAIRAELAGRRNVELHFERFAAPPVVDGRPFSVRIAST
Mflv_3426|M.gilvum_PYR-GCK TSVYACGPAPMLTAVRDALVGRDDVELHFERFAAPPVVDGRPFR--VAAA
Mvan_3180|M.vanbaalenii_PYR-1 TAVYACGPAPMLTAVRAALVGRDDVELHFERFAAPPVTDGRPFQATVAST
MAB_4020c|M.abscessus_ATCC_199 LAVYCCGPAPMLNVVRDAVAECDGAELHFERFSAPPIEHGVPFQVQLGIG
MMAR_1933|M.marinum_M TAVYVCGPPPMLEAVRAARAEYPCAPLHYERFSPPPVVDGVPFELELART
MUL_2159|M.ulcerans_Agy99 TAVYVCGPPPMLEAVRAARAEYPCAPLHYERFSPPPVVDGVPFELELART
Mb2798c|M.bovis_AF2122/97 TAVYVCGPPGMLEAVRVARNQHADAPLHYERFSPPPVVDGVPFELELARS
Rv2776c|M.tuberculosis_H37Rv TAVYVCGPPGMLEAVRVARNQHADAPLHYERFSPPPVVDGVPFELELARS
MAV_4119|M.avium_104 TAVYVCGPTAMLEAVRTARDEHADAPLHYERFGPPPVVDGVPFELELARS
:** ***. ** :* . **:***..**: .* * :.
MSMEG_3676|M.smegmatis_MC2_155 GQTVPVAADETLLAALNRAGVSAPYSCQQGFCGTCRVRALPRDGDGAVQH
TH_3804|M.thermoresistible__bu GAEVPVGADETLLAALGRAGVQAPYSCQQGFCGTCRTRVL----DGTVEH
Mflv_3426|M.gilvum_PYR-GCK GVTVDVGVDETLLAALGRAGVTTPYSCRQGFCGTCRTRVL----SGEVDH
Mvan_3180|M.vanbaalenii_PYR-1 GQQVDVGADETLLAALRRAGVDASYSCQQGFCGTCRTRVL----AGSVEH
MAB_4020c|M.abscessus_ATCC_199 GPVLEVGADQSALAAVLTSRPGTPHSCRQGFCGTCKVKVL----AGRPDH
MMAR_1933|M.marinum_M GEVLSIPANRTALDVMLDRDPTTAYSCQQGFCGTCKVKVL----AGQVDH
MUL_2159|M.ulcerans_Agy99 GEVLSIPANRTALDVMLDRDPTTAYSCQQGFCGTCKVKVL----AGQVDH
Mb2798c|M.bovis_AF2122/97 RRVLRVPANRSALDVMLDWDPTTAYSCQQGFCGTCKVRVL----AGQVDR
Rv2776c|M.tuberculosis_H37Rv RRVLRVPANRSALDVMLDWDPTTAYSCQQGFCGTCKVRVL----AGQVDR
MAV_4119|M.avium_104 RRVLTVPGNRSALDVMLDRDPATPYSCQQGFCGTCKVKVL----AGQVDR
: : :.: * .: :.:**:*******:.:.* * ::
MSMEG_3676|M.smegmatis_MC2_155 RDTLLTEPERAAGYLLTCVSRAGNGQRLTIEL
TH_3804|M.thermoresistible__bu RDRLLTEPERAAGLMLTCVSRAPAGERLTLDL
Mflv_3426|M.gilvum_PYR-GCK RDTLLTDAERAAGLMLPCVSRAAEGTVLALDL
Mvan_3180|M.vanbaalenii_PYR-1 RDTLLTGPERAAGLMLTCVSRAGDGSGLTLDL
MAB_4020c|M.abscessus_ATCC_199 RDSLLTETQRAEGQMLLCVSRAD-GERLVLDI
MMAR_1933|M.marinum_M RGRAAQGDD----EMLVCVSRGH-SARVVIDA
MUL_2159|M.ulcerans_Agy99 RGRAAQGDD----EMLVCVSRGH-SARVVIDA
Mb2798c|M.bovis_AF2122/97 RGRIIEGDN----EMLVCVSRAV-SGRVVIDA
Rv2776c|M.tuberculosis_H37Rv RGRIIEGDN----EMLVCVSRAV-SGRVVIDA
MAV_4119|M.avium_104 RGRTVEGDD----EMLVCVSRAK-NDRLVIDA
*. : :* ****. . :.::