For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVTSIHSRSADCRSFAIASTALAVACRTEGRGAQKCVTVSVRGEVDALNAKHFAHTVREMAGGSAALVL DLTEVSFLAFDGASALYAISAHLAREDVTWCVVPSPAVSRVVKLCDPEGLIPLQDIASVRGVKPA
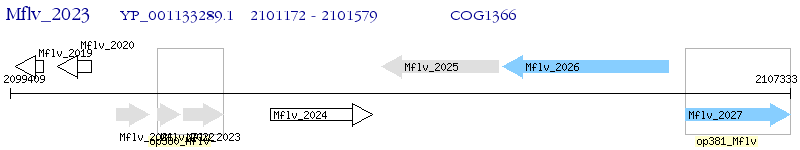
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2023 | - | - | 100% (135) | anti-sigma-factor antagonist |
| M. gilvum PYR-GCK | Mflv_3879 | - | 3e-17 | 46.59% (88) | anti-sigma-factor antagonist |
| M. gilvum PYR-GCK | Mflv_4788 | - | 4e-13 | 38.10% (84) | anti-sigma-factor antagonist |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0529c | - | 4e-08 | 37.68% (69) | hypothetical protein Mb0529c |
| M. tuberculosis H37Rv | Rv0516c | - | 4e-08 | 37.68% (69) | hypothetical protein Rv0516c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2512 | - | 2e-10 | 34.09% (88) | hypothetical protein MAB_2512 |
| M. marinum M | MMAR_0214 | - | 1e-07 | 32.97% (91) | hypothetical protein MMAR_0214 |
| M. avium 104 | MAV_4257 | - | 3e-14 | 38.10% (84) | STAS domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1786 | - | 7e-12 | 38.89% (90) | STAS domain-containing protein |
| M. thermoresistible (build 8) | TH_2508 | - | 2e-14 | 41.86% (86) | PUTATIVE putative anti-sigma-factor antagonist |
| M. ulcerans Agy99 | MUL_0605 | - | 3e-06 | 33.33% (69) | hypothetical protein MUL_0605 |
| M. vanbaalenii PYR-1 | Mvan_4700 | - | 5e-54 | 76.30% (135) | anti-sigma-factor antagonist |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2023|M.gilvum_PYR-GCK ----MTVTSIHSRSADCRSFAIASTALAVACRTEGRGAQKCVTVSVRGEV
Mvan_4700|M.vanbaalenii_PYR-1 ----MTVTSIRSRSADCRRFSIASTSLTFACRTEGAGSATRATVSVWGEV
MSMEG_1786|M.smegmatis_MC2_155 ----MPTISVAKRSSPHSG-SHAPQRAHFTTEQINTAT---AVVTVHGDL
TH_2508|M.thermoresistible__bu ------MILPVPRPSPGGGEAVTCHAADFATRWVNSAT---VVIRVDGEL
MAV_4257|M.avium_104 -------MSVAQSWQ-------QSRTAQFTARWGLTG----TLITVDGEL
Mb0529c|M.bovis_AF2122/97 ---MTTTIPTSKSACSVTTRPGNAAVDYGGAQIRAYLHHLATVVTIRGEI
Rv0516c|M.tuberculosis_H37Rv ---MTTTIPTSKSACSVTTRPGNAAVDYGGAQIRAYLHHLATVVTIRGEI
MUL_0605|M.ulcerans_Agy99 -------MGIAITASSFADRQKNGTFECSGAVVRAHCRHLATIVTIRGEI
MAB_2512|M.abscessus_ATCC_1997 MSVIDQNASRFAARAKTGVVSLFSSADDTARFTAHWHRPSAVLVRATGEI
MMAR_0214|M.marinum_M ---------MVEQSGD--------PTPPTAFGIEKSRVGEAVVVTVSGEL
. : *::
Mflv_2023|M.gilvum_PYR-GCK DALNAKHFAHTVRE-MAGGSAALVLDLTEVSFLAFDGASALYAISAHLAR
Mvan_4700|M.vanbaalenii_PYR-1 DAANAKHFAHTVRE-VAEGNAALILDLSEVNFMAFDGASALYAISAQLAR
MSMEG_1786|M.smegmatis_MC2_155 DASNAGLLTDYALK-ALTRNAKLVLDLSDVAFFGAACFTALHTLNVRCAG
TH_2508|M.thermoresistible__bu DAANSPQLVDYALR-HAGRIKYLVADLTGVEFFGIAGFSAAHDINGRCAH
MAV_4257|M.avium_104 DAANADQLAAYVQQ-SVNRSRRVILDLRGLNFIGTAGFSALHRINVICSA
Mb0529c|M.bovis_AF2122/97 DAANVEQISEHVRR-FSLGTNPMVLDLSELSHFSGAGISLLCILDEDCRA
Rv0516c|M.tuberculosis_H37Rv DAANVEQISEHVRR-FSLGTNPMVLDLSELSHFSGAGISLLCILDEDCRA
MUL_0605|M.ulcerans_Agy99 DAVNVDQVTERIQH-FIHGTNPVVLDVGDVSHFSPAGISLLAAIDEECHA
MAB_2512|M.abscessus_ATCC_1997 DAVNANHLAEYATR-YLDGHRTLVLDLQGLRFFGTDGLVALDRVRRHCKV
MMAR_0214|M.marinum_M DMLTAPRLAEMLRTSLIEAPATLIVDLSKVDFLASAGMTVLVSAKAQAVA
* . . :: *: : .:.
Mflv_2023|M.gilvum_PYR-GCK EDVTWCVVPSPAVSRVVK----LCDPEGLIPLQDIASVRG---------V
Mvan_4700|M.vanbaalenii_PYR-1 EGVTWCVIASPAVSRVLE----LCDPEGLIPATTIPSVRG---------A
MSMEG_1786|M.smegmatis_MC2_155 ADAEWVVVPSKAVTRVLR----ICDPDSGLTTAPDVVSALSR------KS
TH_2508|M.thermoresistible__bu AKLTWTLVPSPAVVRLLQ----ICDPDGTLPLNTTVPDSVWD------NG
MAV_4257|M.avium_104 AQTSWAMAPSPAVARLLR----LCDPDGTLPVTTPKAEPLLEPRRANGDE
Mb0529c|M.bovis_AF2122/97 AGVQWALVASPAVVEQLGGRCDQGEHESMFPMARSVHKALHDLADAI-DR
Rv0516c|M.tuberculosis_H37Rv AGVQWALVASPAVVEQLGGRCDQGEHESMFPMARSVHKALHDLADAI-DR
MUL_0605|M.ulcerans_Agy99 AGAEWTLVAGPAVVELLG-----EDMATMFPVAHSVHEALRNLAEAN-SS
MAB_2512|M.abscessus_ATCC_1997 QGIDWTVIPGDAVTKLLT----LSGDMAAYPVNDAIPDVWKKLAQ-----
MMAR_0214|M.marinum_M PTRFVVVADGPATSRPIR----LMGIDAMLPLYDALDDALNDISDS----
: . *. . : .
Mflv_2023|M.gilvum_PYR-GCK KPA----------
Mvan_4700|M.vanbaalenii_PYR-1 KPA----------
MSMEG_1786|M.smegmatis_MC2_155 DQR-PLQLVENT-
TH_2508|M.thermoresistible__bu GSGGLFQLIPEAG
MAV_4257|M.avium_104 SPGPLLQLVTKPR
Mb0529c|M.bovis_AF2122/97 RRQLVLPLISRSA
Rv0516c|M.tuberculosis_H37Rv RRQLVLPLISRSA
MUL_0605|M.ulcerans_Agy99 RRQLMLSLVRKTA
MAB_2512|M.abscessus_ATCC_1997 -------------
MMAR_0214|M.marinum_M -------------