For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RGLTGKTFIIAGGATGIGAGTAKRLADEGANVVVGDIAIAAATATAEKLCAVGARAIAVEFDLADEFSVR QLVEHTITEFGSVDGLFNVGADLSPETNGTDLTILDTGLDIWQRTLDINLLGYVRTIRAVLPHLLEQGSG SIVNTSSGGSLGTDPMHVAYNASKAAVNQLTRHVANNYGAKGIRSNCVMPGLVWGETQEKANLEPLREMF VAAARTTRVGRPSDLASITAFLLSDEAEWINGQCWYIGGASHMRQ*
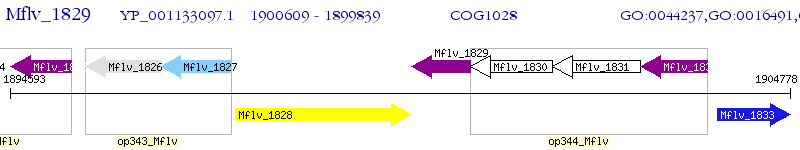
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1829 | - | - | 100% (256) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_3483 | - | 2e-57 | 46.09% (256) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_2001 | - | 3e-36 | 34.82% (247) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0792 | - | 2e-32 | 35.20% (250) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0769 | - | 6e-33 | 35.60% (250) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 2e-16 | 33.33% (195) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1219 | - | 2e-28 | 32.40% (250) | short chain dehydrogenase |
| M. marinum M | MMAR_0271 | - | 3e-35 | 35.68% (241) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_3097 | - | 1e-104 | 73.05% (256) | NAD dependent epimerase/dehydratase family protein |
| M. smegmatis MC2 155 | MSMEG_2598 | - | 5e-33 | 36.82% (258) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3037 | - | 7e-32 | 34.00% (250) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4844 | - | 7e-34 | 34.85% (241) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_5156 | - | 2e-30 | 32.80% (250) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1829|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3097|M.avium_104 --------------------------------------------------
MMAR_0271|M.marinum_M --------------------------------------------------
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1829|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3097|M.avium_104 --------------------------------------------------
MMAR_0271|M.marinum_M --------------------------------------------------
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1829|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3097|M.avium_104 --------------------------------------------------
MMAR_0271|M.marinum_M --------------------------------------------------
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1829|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3097|M.avium_104 --------------------------------------------------
MMAR_0271|M.marinum_M --------------------------------------------------
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1829|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3097|M.avium_104 --------------------------------------------------
MMAR_0271|M.marinum_M --------------------------------------------------
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1829|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3097|M.avium_104 --------------------------------------------------
MMAR_0271|M.marinum_M --------------------------------------------------
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb0792|M.bovis_AF2122/97 ------------------------MFDSKVAIVTGAAQGIGQAYAQALAR
Rv0769|M.tuberculosis_H37Rv ------------------------MFDSKVAIVTGAAQGIGQAYAQALAR
MAB_1219|M.abscessus_ATCC_1997 ------------------MG----EFDNKVAIVTGAAQGIGEAYAHALAR
TH_3037|M.thermoresistible__bu ------------------MG----QFDDKVAIVTGAGGGIGQAYAEALAR
Mvan_5156|M.vanbaalenii_PYR-1 ------------------MGLYGDQFKEKVAIVTGAGGGIGQAYAEALAR
Mflv_1829|M.gilvum_PYR-GCK ----------------------MRGLTGKTFIIAGGATGIGAGTAKRLAD
MAV_3097|M.avium_104 ----------------------MRGLQGKTFIVAGGATGIGAATAERLAS
MMAR_0271|M.marinum_M ---------------------MTAELENKVAIITGACGGIGLETSRVLAR
MUL_4844|M.ulcerans_Agy99 ---------------------MTAELENKVAIITGACGGIGLETSRVPAC
MSMEG_2598|M.smegmatis_MC2_155 -------------------MDLTQRLAGKVAVITGGASGIGLATGRRLRA
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
: . ::*. *** .
Mb0792|M.bovis_AF2122/97 EGASVVVADINADGAAAVAKQIVADGGTVIHVPVDVSDEDSAKAMVDRAV
Rv0769|M.tuberculosis_H37Rv EGASVVVADINADGAAAVAKQIVADGGTAIHVPVDVSDEDSAKAMVDRAV
MAB_1219|M.abscessus_ATCC_1997 EGAAVVVADINAEMGEGVAKQIVADGGRAIFAGVDVSDPDSAIAMADKAV
TH_3037|M.thermoresistible__bu EGAAVVVADINTEGAQKVADGIKGEGGDALAVPVDVSDPESAEEMAAQTV
Mvan_5156|M.vanbaalenii_PYR-1 EGAAVVVADINTEGAQKVADGIKGEGGNALAVRVDVSDPESAKGMAAQTL
Mflv_1829|M.gilvum_PYR-GCK EGANVVVGDIAIAAATATAEKLCAVGARAIAVEFDLADEFSVRQLVEHTI
MAV_3097|M.avium_104 EGAAVTVGDINIEGANATVGRITQSGGRAIAVEFDLADDASVRNLVQKTI
MMAR_0271|M.marinum_M AGARVVLADLPETDL---AGAAASVGRGAVHHVVDLTNEVSVRALIDFTI
MUL_4844|M.ulcerans_Agy99 AGARVVLADLPETDL---AGAAASVGRGAVHHVVDLTNEVSVRALIDFTI
MSMEG_2598|M.smegmatis_MC2_155 EGATVVVGDIDPT-----TGKAAADELEGLFVPVDVSEQEAVDNLFDTAA
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
** *.:.*: . .*::: :. : .
Mb0792|M.bovis_AF2122/97 GAFGGIDYLVNNAAIYGGMK---LDLLLTVPLDYYKKFMSVNHDGVLVCT
Rv0769|M.tuberculosis_H37Rv GAFGGIDYLVNNAAIYGGMK---LDLLLTVPLDYYKKFMSVNHDGVLVCT
MAB_1219|M.abscessus_ATCC_1997 SEFGGIDYLVNNAAIYGGMK---LDLLLSVPWDYYKKFMSVNQDGALVCT
TH_3037|M.thermoresistible__bu SEFGGIDYLVNNAAIFGGMK---LDFLITVDWDYYKRFMSVNLDGALVCT
Mvan_5156|M.vanbaalenii_PYR-1 SEFGGIDYLVNNAAIFGGMK---LDFLITVDWDYYKKFMSVNMDGALVCT
Mflv_1829|M.gilvum_PYR-GCK TEFGSVDGLFNVGADLSPETNGTDLTILDTGLDIWQRTLDINLLGYVRTI
MAV_3097|M.avium_104 GEFGALHGLHNVGADLSAENLGRDTTLLDTDFDVWRRTLDVNLLGYVRTS
MMAR_0271|M.marinum_M DTFGRLDIVDNNAAHSDPA----DMLVTQMTVDVWDDTFTVNARGTMLMC
MUL_4844|M.ulcerans_Agy99 DTFGRLDIVDNNAAHSDPA----DMLVSQMTVDVWDDTFTVNARGTMLMC
MSMEG_2598|M.smegmatis_MC2_155 STFGRVDIAFNNAGISPPE----DDLIENTDLPAWQRVQDINLKSVYLSC
MLBr_00862|M.leprae_Br4923 AKHGVPDIVVNNAGIGQAGR------FLDTPPEEFDRVLAVNLGGVVNCC
.* . * .. . : :* .
Mb0792|M.bovis_AF2122/97 RAVYKHMAKRGGGAIVNQSS----TAAWLYSNFYGLAKVGVNGLTQQLAR
Rv0769|M.tuberculosis_H37Rv RAVYKHMAKRGGGAIVNQSS----TAAWLYSNFYGLAKVGVNGLTQQLAR
MAB_1219|M.abscessus_ATCC_1997 RAVYKHIAQRGGGAIVNQSS----TAAWLYSGFYGLAKVGINGLTQQLSR
TH_3037|M.thermoresistible__bu RAVYKKMAKRGGGAIVNQSS----TAAWLYSNYYGLAKAGLNSLTQQLAT
Mvan_5156|M.vanbaalenii_PYR-1 RAVYRKMAKRGGGAIVNQSS----TAAWLYSNFYGLAKVGINGLTQQLAT
Mflv_1829|M.gilvum_PYR-GCK RAVLPHLLEQGSGSIVNTSSGGSLGTDPMH-VAYNASKAAVNQLTRHVAN
MAV_3097|M.avium_104 RAVLAHLLQQGSGSIVNTSSGGSLGTDPMH-VAYNAAKAAVNQLTRHIAN
MMAR_0271|M.marinum_M KYAIPRLISAGGGAIVNISSATAHAAYDMS-TAYACTKAAIETLTRYVAT
MUL_4844|M.ulcerans_Agy99 KYAIPRLISAGGGAIVNISSATAHAAYDMS-TAYSCTKAAIETLTRYVAT
MSMEG_2598|M.smegmatis_MC2_155 RAALRHMVPAGKGSIINTASFVAVMGSATSQISYTASKGGVLAMSRELGV
MLBr_00862|M.leprae_Br4923 RAFGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRA
: :: * *. : * * :* . :: :
Mb0792|M.bovis_AF2122/97 ELGGMKIRINAIAPGPIDTEATRTVTPAE--LVKNMVQT--------IPL
Rv0769|M.tuberculosis_H37Rv ELGGMKIRINAIAPGPIDTEATRTVTPAE--LVKNMVQT--------IPL
MAB_1219|M.abscessus_ATCC_1997 ELGGQKIRINAIAPGPTDTEATRGTVPEQ--FRSELVKN--------IPL
TH_3037|M.thermoresistible__bu ELGGQNIRVNAIAPGPIDTEANRSTTPKE--MVDDIVKR--------IPL
Mvan_5156|M.vanbaalenii_PYR-1 ELGGQNIRVNAIAPGPIDTEANRTTTPQE--MVADIVKG--------IPL
Mflv_1829|M.gilvum_PYR-GCK NYGAKGIRSNCVMPGLVWGETQEKANLEP--LREMFVAA--------ART
MAV_3097|M.avium_104 NWGAQGVRCNGVMPGLVMGETQKQQNDIQ--LQQMFLQA--------AKV
MMAR_0271|M.marinum_M QYGRHGVRCNAIAPGLVRTPRLEVGLPQP--IVDIFATH--------HLA
MUL_4844|M.ulcerans_Agy99 QYGRHGVRCNAIAPGLVRTPRLEVGLPQP--IVDIFATH--------HLA
MSMEG_2598|M.smegmatis_MC2_155 QYARQGIRVNALCPGPVNTPLLQELFAKDPERAARRLVH--------IPL
MLBr_00862|M.leprae_Br4923 ELDAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRL
: : . : ** .
Mb0792|M.bovis_AF2122/97 SRMGTPEDLVGMCLFLLSDSASWITG-QIFNVDG-------------GQI
Rv0769|M.tuberculosis_H37Rv SRMGTPEDLVGMCLFLLSDSASWITG-QIFNVDG-------------GQI
MAB_1219|M.abscessus_ATCC_1997 SRMGTVDDMVGMCLFLLSDKASWITG-QVFNVDG-------------GQI
TH_3037|M.thermoresistible__bu SRLGEPEDLVGMCLFLLSDQAKWITG-QIFNVDG-------------GQI
Mvan_5156|M.vanbaalenii_PYR-1 SRMGEPEDLVGMLLFLLSDQAKWITG-QIFNVDG-------------GQI
Mflv_1829|M.gilvum_PYR-GCK TRVGRPSDLASITAFLLSDEAEWING-QCWYIGG-------------ASH
MAV_3097|M.avium_104 TRLGEPRDIAAITAFLLSDEAEWING-QVWYIGG-------------ASH
MMAR_0271|M.marinum_M GRIGEPHEIAELVCFLASDRAAFITG-QVIAADSGLLAHLPGLPQIRASV
MUL_4844|M.ulcerans_Agy99 GLIGEPHEIAELVCFLASDRAAFITG-QVIAADSGLLAHLPGLPQIRASV
MSMEG_2598|M.smegmatis_MC2_155 GRFAEPEELAAAVAFLASDDASFITG-STFLVDGGISS---------AYV
MLBr_00862|M.leprae_Br4923 RRYGPDKVADTILSAIKKNKPIRPVAPEAYALYG--------ISRVLPQV
. : .: . . . .
Mb0792|M.bovis_AF2122/97 IRS-------
Rv0769|M.tuberculosis_H37Rv IRS-------
MAB_1219|M.abscessus_ATCC_1997 IRS-------
TH_3037|M.thermoresistible__bu IR--------
Mvan_5156|M.vanbaalenii_PYR-1 IRS-------
Mflv_1829|M.gilvum_PYR-GCK MRQ-------
MAV_3097|M.avium_104 MRQ-------
MMAR_0271|M.marinum_M AELQSQPS--
MUL_4844|M.ulcerans_Agy99 AELQSQPS--
MSMEG_2598|M.smegmatis_MC2_155 TPL-------
MLBr_00862|M.leprae_Br4923 LRSTARIRVI