For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAVLSHGSPARFEPANASTWRDPWPMYRALRDHDPVHHVVPADPQHDYYVLSRHADIWAAARDHETFSSA QGLTVNYGELDLIGLADNPPMVMQDPPGHTEFRRLVARGFTPRQVEAVEPKVREFVVERIERLRADGGGD IVAELFKPLPSMVVAHYLGVPDEDRGKFDGWTDAIVAANTTDGGIAGALGTLGDALGEMMGYFTALIERR RSEPADDTVSHLVAAGVGADGDISGVLSILAFTFTMVTGGNDTTTGMLGGSVQLLHQRPDQRRLLADNPN LIPDAVDEFLRLTSPVQMLGRTVTRDVTLHDVTIPEGRRVMFLYGSGNRDERHFGDDAGELDVTRRPRNI LTFSHGAHHCLGAAAARMQSRVALTELLTRIPNFEVDESGIVWAGGSYVRRPLSVPFTVTS*
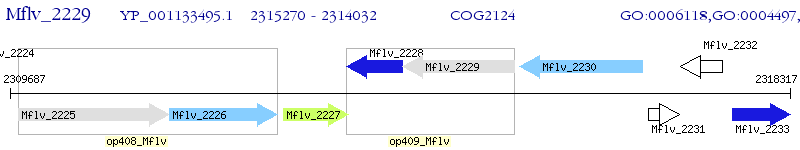
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2229 | - | - | 100% (412) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_1321 | - | 8e-63 | 39.89% (371) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_2477 | - | 1e-59 | 35.99% (389) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0789c | cyp123 | 4e-59 | 36.59% (399) | cytochrome P450 123 |
| M. tuberculosis H37Rv | Rv1256c | cyp130 | 0.0 | 77.72% (413) | cytochrome P450 130 CYP130 |
| M. leprae Br4923 | MLBr_02088 | - | 6e-39 | 29.37% (378) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_1406c | - | 1e-172 | 71.50% (414) | cytochrome P450 |
| M. marinum M | MMAR_4184 | cyp130A4 | 0.0 | 79.90% (413) | cytochrome P450 130A4 Cyp130A4 |
| M. avium 104 | MAV_1404 | - | 0.0 | 80.93% (409) | putative cytochrome P450 130 |
| M. smegmatis MC2 155 | MSMEG_5038 | - | 0.0 | 78.97% (409) | putative cytochrome P450 123 |
| M. thermoresistible (build 8) | TH_3609 | - | 1e-178 | 76.67% (403) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_0324 | cyp189A7 | 8e-65 | 35.38% (407) | cytochrome P450 189A7 Cyp189A7 |
| M. vanbaalenii PYR-1 | Mvan_4465 | - | 0.0 | 90.05% (412) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2229|M.gilvum_PYR-GCK --------------------------MTAVLSHGSPARFEPANASTWRDP
Mvan_4465|M.vanbaalenii_PYR-1 --------------------------MTAVLSHGSPVHFALADASTWADP
Rv1256c|M.tuberculosis_H37Rv --------------------------MTSVMSH----EFQLATAETWPNP
MMAR_4184|M.marinum_M --------------------------MSVDVSHRAPVAFQLATAATWPDP
MAV_1404|M.avium_104 ------------------------------MSHNVPVAFELPNADTWADP
MSMEG_5038|M.smegmatis_MC2_155 --------------------------MGADLSHP-AVAFELANADTWADP
TH_3609|M.thermoresistible__bu ------------------------------VSQT-PVRFTLSTAATWSDP
MAB_1406c|M.abscessus_ATCC_199 --------------------------MSVDVSHTVAPRYVPGTADNWTNP
MUL_0324|M.ulcerans_Agy99 ----------------------------MTVTAASDVYFDPYDVELNADP
Mb0789c|M.bovis_AF2122/97 ---------------------------MTVRVGDPELVLDPYDYDFHEDP
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
:*
Mflv_2229|M.gilvum_PYR-GCK WPMYRALRDHDPVHHVVPAD-PQHDYYVLSRHADIWAAARDHETFSSAQG
Mvan_4465|M.vanbaalenii_PYR-1 WPMYRALRDHDPVHHVVPEDRPNHDYYVLSRHADIWAAARDHQTFSSALG
Rv1256c|M.tuberculosis_H37Rv WPMYRALRDHDPVHHVVPPQRPEYDYYVLSRHADVWSAARDHQTFSSAQG
MMAR_4184|M.marinum_M WPMYRALRDQDPVHHVIPAHQPDQDYYVLSRHADVWAAARDHQTFSSAQG
MAV_1404|M.avium_104 WPMYRALRDHDPVHHVVPPKRPEHDYYVLSRHADVWAAARDHETFSSAKG
MSMEG_5038|M.smegmatis_MC2_155 WDMYTALRDHDPVHHVVPPERPHHDYFVLSRHADIWAAARDHETYSSAQG
TH_3609|M.thermoresistible__bu WPMYAALRDHDPVHHVVPEHRPEHDYYVLSRHADVWRAARDHETFSSARG
MAB_1406c|M.abscessus_ATCC_199 WPMYAALRDHDPVHHVVPEYTPDQDYYVLTRHADVYEAARDWETFSSARG
MUL_0324|M.ulcerans_Agy99 YPMFRRLREEAPLYYN-----AQHGFYALSRFADVDRAIVDYQTFSSARG
Mb0789c|M.bovis_AF2122/97 YPYYRRLRDEAPLYRN-----EERNFWAVSRHHDVLQGFRDSTALSNAYG
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLP------GMPLTVFSSFSDCDEALRHPLSASDRLK
: : * : *: ..: . * . . : *.
Mflv_2229|M.gilvum_PYR-GCK LTVNYGELDLIG---LADNPPMVMQDPPGHTEFRRLVARGFTPRQVEAVE
Mvan_4465|M.vanbaalenii_PYR-1 LTVNYGELDLIG---LADNPPMVMQDPPVHTEFRKLVARGFTPRQVEAVE
Rv1256c|M.tuberculosis_H37Rv LTVNYGELEMIG---LHDTPPMVMQDPPVHTEFRKLVSRGFTPRQVETVE
MMAR_4184|M.marinum_M LTVNYGDLEMIG---LQDNPPMVMQDPPAHTEFRKLVSRGFTPRQVEAVE
MAV_1404|M.avium_104 LTVNYDDLELIG---LQDNPPFVMQDPPVHTEFRKLVSRGFTPRQVEAVE
MSMEG_5038|M.smegmatis_MC2_155 LTVNYGELELIG---LADNPPFVMQDPPTHTEFRKLVSRGFTPRQVEAVE
TH_3609|M.thermoresistible__bu LTVNYDELELIG---LADNPPFVMQDPPVHTEFRKLVSRGFTPRQVTAVE
MAB_1406c|M.abscessus_ATCC_199 LTVTYGDLEKTG---MGDNPPMVMQDPPTHTEFRKLVSRGFTPRQVTAVE
MUL_0324|M.ulcerans_Agy99 AILEFIKANIDI---PAG--VLVFEDPPVHDVHRKLLSRMFTPRKINDLE
Mb0789c|M.bovis_AF2122/97 VSLDPSSRTSEA---YRVMSMLAMDDP-AHLRMRTLVSKGFTPRRIRELE
MLBr_02088|M.leprae_Br4923 ATLAQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVVQALE
: : : ** * * *::: *:*: : :*
Mflv_2229|M.gilvum_PYR-GCK PKVREFVVERIERLRADGGGDIVAELFKPLPSMVVAHYLGVPDEDRGKFD
Mvan_4465|M.vanbaalenii_PYR-1 PKVHDFVVDRIEHLRANGGGDIVAELFKPLPSMVVAHYLGVPDEDRGKFD
Rv1256c|M.tuberculosis_H37Rv PTVRKFVVERLEKLRANGGGDIVTELFKPLPSMVVAHYLGVPEEDWTQFD
MMAR_4184|M.marinum_M PKVRDFVVERIERLRAQGGGDIVTELFKPLPSMVVAHYLGVPEQDRTQFD
MAV_1404|M.avium_104 PKVRDFVVERIERLRAAGGGDIVAELFKPLPSMVVAHYLGVPEEDRAQFD
MSMEG_5038|M.smegmatis_MC2_155 PQVREFVVERLERLRAEGGGDIVAELFKPLPSMVVAHYLGVPEADRAQFD
TH_3609|M.thermoresistible__bu PAVRRFVVERIERLRADGGGDIVAELFKPLPSMVVAHYLGVPESDRAQFD
MAB_1406c|M.abscessus_ATCC_199 PKVREFVVQRLERLKERGEGDIVVELFKPLPSMVVAHYLGVPEEDRTQFD
MUL_0324|M.ulcerans_Agy99 PKIREFCARSLDPLIGSGRFDFVADLGAQMPMRVIGMLLGVPEEDQEAAR
Mb0789c|M.bovis_AF2122/97 PQVLELARIHLDSALQTESFDFVAEFAGKLPMDVISELIGVPDTDRARIR
MLBr_02088|M.leprae_Br4923 GDIAALVDSLLDKGAAAGQFDVIADLAFPLAVAVICRLLGVPYEDAPEFG
: : :: *.:.:: :. *: :*** *
Mflv_2229|M.gilvum_PYR-GCK GWTDAIVAANT----TDGGIAGALGTLGDALGEMMGYFTALIERRRSEPA
Mvan_4465|M.vanbaalenii_PYR-1 GWTDAIVAANT----SAGGLGGALDTLGDALGEMMSYFTALIERRRTEPE
Rv1256c|M.tuberculosis_H37Rv GWTQAIVAANA----VDGATTGAL----DAVGSMMAYFTGLIERRRTEPA
MMAR_4184|M.marinum_M GWTQAIVAANT----AEGGIAGALATVGDAVGSMMAYFTSLIERRRAKPE
MAV_1404|M.avium_104 GWTEAIVAANT----ADGGVAGALGSAGDAVTSMMAYFTGLIERRRTDPE
MSMEG_5038|M.smegmatis_MC2_155 GWTEAIVAANS----EAGSALGAG----DAVMSMTGYFAELIERRRREPG
TH_3609|M.thermoresistible__bu RWTDAIVAANT----GADGVMGAG----AAVGEMMAYFTELIEYRRREPA
MAB_1406c|M.abscessus_ATCC_199 AWTDGIVAAASG---GAIDIEAMQGEVAQTIGELMMYFTGLIERRRAEPE
MUL_0324|M.ulcerans_Agy99 DFATAQLRTEAG---KPMKASADG-------MVSGDFFAHYIDWRAEHPS
Mb0789c|M.bovis_AF2122/97 ALADAVLHRED-------GVADVPPPAMAASIELMRYYADLIAEFRRRPA
MLBr_02088|M.leprae_Br4923 RVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRGTPG
: : : : *
Mflv_2229|M.gilvum_PYR-GCK DDTVSHLVAAGVGADGDISGVLS---ILAFTFTMVTGGNDTTTGMLGGSV
Mvan_4465|M.vanbaalenii_PYR-1 DDTISHLVAAGVGADGDISGVLS---ILAFTFTMVTGGNDTTTGMLGGSV
Rv1256c|M.tuberculosis_H37Rv DDAISHLVAAGVGADGDTAGTLS---ILAFTFTMVTGGNDTVTGMLGGSM
MMAR_4184|M.marinum_M DDTISHLVAAGVGADGDIAGTLS---ILAFTFTMVTGGNDTVTGMLGGSM
MAV_1404|M.avium_104 DDTISHLVSAGVGADGDIAGTLS---VLAFTFTMVTGGNDTTTGMLGGSM
MSMEG_5038|M.smegmatis_MC2_155 DDTISHLVAAGA--DDEAVDVLS---ILAFTFTMVTGGNDTTTGMLGGAV
TH_3609|M.thermoresistible__bu DDTISHLVAAG-------VDPLP---VLAFTFTMVTGGNDTTTGMLGGTM
MAB_1406c|M.abscessus_ATCC_199 DDTVSHLVAAGVGADGDISGTLQ---ILGFAFTMVTGGNDTTTGMLGGAI
MUL_0324|M.ulcerans_Agy99 SDIMTELLNAEFEDETGTVRRLRRDELLTYVSVVSGAGNETTTRLIGWAG
Mb0789c|M.bovis_AF2122/97 NNLTSALLAAELD-----GDRLSDQEIMAFLFLMVIAGNETTTKLLANAV
MLBr_02088|M.leprae_Br4923 EDLISRLIELDES-----GDQLTEEEIIATCGLLLVAGHETTVNLIANAV
.: : *: * :: : .*::*.. ::. :
Mflv_2229|M.gilvum_PYR-GCK QLLHQRPDQRRLLADNPNLIPDAVDEFLRLTSPVQMLGRTVTRDVTLHDV
Mvan_4465|M.vanbaalenii_PYR-1 QLLHRRPDQRTMLAENPDLIPDAVDEFLRLTSPVQMLGRTVTHDVTIGDV
Rv1256c|M.tuberculosis_H37Rv PLLHRRPDQRRLLLDDPEGIPDAVEELLRLTSPVQGLARTTTRDVTIGDT
MMAR_4184|M.marinum_M PLLHRRPDQRRLLTDNPELITDAVEELLRLTSPVQGLARTVTHDVTIGNV
MAV_1404|M.avium_104 PLLHQRPDQRQRLVDEPELIPDAVEELLRLTSPVQGLARTTTRDVTIGRT
MSMEG_5038|M.smegmatis_MC2_155 QLLQQHPDQRRVLTDDPDLIPDAVDEFLRLTSPVQGLARTATRDVTIGDT
TH_3609|M.thermoresistible__bu QLLHRRPDQRRLLVERPELIADAVDEFLRLTSPVQGLARTSTRDVTIGDT
MAB_1406c|M.abscessus_ATCC_199 QLLHQNPAQRQKLIDDPSLIPGAVEEFLRLTSPVQGLARTATRDVIIGDT
MUL_0324|M.ulcerans_Agy99 KVLAEHPDQRRALVEDPSLIPQAVEELLRYEAPAPHVARYVTRDLRYYGQ
Mb0789c|M.bovis_AF2122/97 YWAAHHPGQLARVFADHSRIPMWVEETLRYDTSSQILARTVAHDLTLYDT
MLBr_02088|M.leprae_Br4923 LAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAAKDMTIGQT
..* * : . . *:* ** .. :.* ::*:
Mflv_2229|M.gilvum_PYR-GCK TIPEGRRVMFLYGSGNRDERHFGDDAGELDVTRRPRNILTFSHGAHHCLG
Mvan_4465|M.vanbaalenii_PYR-1 TIPEGRRVMFLYGSGNRDERQYGDDAGELDVTRKPRNILTFSHGAHHCLG
Rv1256c|M.tuberculosis_H37Rv TIPAGRRVLLLYGSANRDERQYGPDAAELDVTRCPRNILTFSHGAHHCLG
MMAR_4184|M.marinum_M TIPAGRRALLLYGSANRDERQYGPDAGELDVRRCPRNILTFSHGAHHCLG
MAV_1404|M.avium_104 TIPAGRRVLLLYGSANRDERQYGPDAGELDVARCPRNILTFSHGAHHCLG
MSMEG_5038|M.smegmatis_MC2_155 TIPAGRKVLLLYGSGNRDEREFGPDAAQLDVRRRPRNILTFSHGAHFCLG
TH_3609|M.thermoresistible__bu TIPAGRKVLLLYASANRDEREFGPDAAELDVTRRPRNILTFSHGAHFCLG
MAB_1406c|M.abscessus_ATCC_199 TIPAGRRALLLYGSANRDEREYGNDAAELDVLRKPRNILTFSHGNHHCLG
MUL_0324|M.ulcerans_Agy99 SVPEGSIMMMLIGAANRDHRQFRPDGDVFNIRREPHQHLTFSVGTHYCLG
Mb0789c|M.bovis_AF2122/97 TIPEGEVLLLLPGSANRDDRVFDDPDDYRIGREIGCKLVSFGSGAHFCLG
MLBr_02088|M.leprae_Br4923 TLTEGDTMVLLLAAANRDPAVYSRP-DEFDPDRPSSRHLAFAVGSHFCLG
::. * ::* .:.*** : . . ::*. * *.***
Mflv_2229|M.gilvum_PYR-GCK AAAARMQSRVALTELLTRIPNFEVDESGIVWAGGSYVRRPLSVPFTVTS-
Mvan_4465|M.vanbaalenii_PYR-1 AAAARMQSRVALTELLTRIPDFEVDESGIAWAGGSYVRRPVSVPFTVTH-
Rv1256c|M.tuberculosis_H37Rv AAAARMQCRVALTELLARCPDFEVAESRIVWSGGSYVRRPLSVPFRVTS-
MMAR_4184|M.marinum_M AAAARMQSRVALTELLARCPDFEVDESRIVWSGGSYVRRPLSLPFEVRS-
MAV_1404|M.avium_104 AAAARMQSRVALTELLARCPDFEVDESGIVWAGGNYVRRPLSVPIRVKS-
MSMEG_5038|M.smegmatis_MC2_155 AAAARMQSRVALTELLTRCPDFTVDLDAVVWAGGSYVRRPLSVPFRAGA-
TH_3609|M.thermoresistible__bu AAAARMQSRVALTELLVRCPEFSVDEDGITWADGGYVRRPLTLPVRIPGQ
MAB_1406c|M.abscessus_ATCC_199 AAAARMQSRIALEELLTRIPDFEVDLDGVTWADGSYVRRPLTVPIRVR--
MUL_0324|M.ulcerans_Agy99 SALARLEGRIALEDILKRFPEWDVDLGNAKLSPTSTVRGWESMPAVFD--
Mb0789c|M.bovis_AF2122/97 AHLARMEARVALGALLRRIRNYEVDDDNVVRVHSSNVRGFAHLPISVQAR
MLBr_02088|M.leprae_Br4923 AALARLEATVTLSAISARFPQVQLAG-ELVYKPNVAMRGMSALPVQV---
: **:: ::* : * : : :* :*
Mflv_2229|M.gilvum_PYR-GCK ----
Mvan_4465|M.vanbaalenii_PYR-1 ----
Rv1256c|M.tuberculosis_H37Rv ----
MMAR_4184|M.marinum_M ----
MAV_1404|M.avium_104 ----
MSMEG_5038|M.smegmatis_MC2_155 ----
TH_3609|M.thermoresistible__bu APRR
MAB_1406c|M.abscessus_ATCC_199 ----
MUL_0324|M.ulcerans_Agy99 ----
Mb0789c|M.bovis_AF2122/97 ----
MLBr_02088|M.leprae_Br4923 ----