For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDLRRTDVATMLLDRAGDQHLGVLTRERDWTWDQVVRESAARASLATRLLDGVPGPFHVGILLANVPDFV FWLGGAALAGATVVGLNPTRGTADLAADIRHADCRLIVTDSVGAQRLRGIGHGVPDDRVLVVDDPEYRRA LEACATEPAVAAGVGASSLFLLLFTSGTTGASKAVKCSQGRLARIAYVATEKFGHHRGDVDYCCMPLFHG NALMALWAPALANGATVCLTPSFSASRFLSDIRYFGATFFTYVGKALAYLMATPEQADDSDNTLTRGFGT EASPEDQAEFRRRFGAELHEGYGSSEGGAVATPDPAAPATALGRPAHEDVVVVDPATLKPCAPAVLDPRG RVTNADDAIGEIVDRRGARDFEGYYRNDAADADRVRNGWYWSGDLGYVDRDGFLYFAGRRGDWIRVDGEN TSALTIERVLRRHPEVIAAAVYGVPDPRSGDQVMAAVEVAEPGSFDINAFVDFLVGQDDLGAKGFPRLLR VSARLPATGSNKVLKRELQTQRWHTDEAVYRWSGRGRPRYRLMTADDRAALDEEFVTHGRAHHGTT*
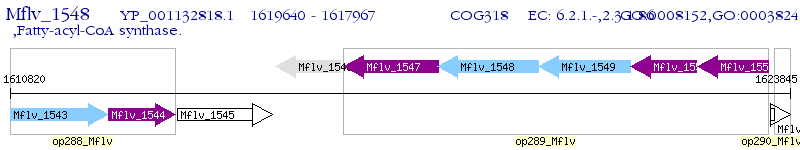
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1548 | - | - | 100% (557) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_2305 | - | e-108 | 43.11% (501) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_1549 | - | e-105 | 44.61% (482) | acyl-CoA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1779c | fadD1 | 1e-109 | 43.14% (503) | acyl-CoA synthetase |
| M. tuberculosis H37Rv | Rv1750c | fadD1 | 1e-109 | 43.14% (503) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_02661 | - | 6e-23 | 25.30% (498) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_3628c | - | 0.0 | 72.53% (546) | acyl-CoA synthetase |
| M. marinum M | MMAR_4994 | fadD17 | 1e-108 | 43.35% (496) | fatty-acid-CoA synthetase FadD17 |
| M. avium 104 | MAV_5257 | - | 1e-108 | 42.57% (505) | acyl-CoA synthetase |
| M. smegmatis MC2 155 | MSMEG_4952 | - | 1e-101 | 41.80% (500) | acyl-CoA synthetase |
| M. thermoresistible (build 8) | TH_1138 | fadD17 | 1e-101 | 41.90% (506) | POSSIBLE FATTY-ACID-CoA SYNTHETASE FADD17 (FATTY-ACID-CoA |
| M. ulcerans Agy99 | MUL_4069 | fadD17 | 1e-107 | 43.35% (496) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_5209 | - | 0.0 | 79.45% (550) | acyl-CoA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1548|M.gilvum_PYR-GCK -------MTDLRRTDVATMLLDRAGDQHLGVLTRERDWTWDQVVRESAAR
Mvan_5209|M.vanbaalenii_PYR-1 --------MTARRDDVAAMLLDRTGDRRPGLRTRERDWTWDEVVHESAVR
MAB_3628c|M.abscessus_ATCC_199 MTAVSVPRAGVPRVDIAAMLLDRVGDSHLGLRTRDRDWTWDQVVDESAAR
MMAR_4994|M.marinum_M ---------MSADPTVPQLLVPLAEIGDRGIYFEDSFTSWADHIGHGAAI
MUL_4069|M.ulcerans_Agy99 ---------MSADPTVPQLLVPLAEIGDRGIYFEDSFTSWADHIGHGAAI
TH_1138|M.thermoresistible__bu -----------MAETVTALLEPLIDVEDRGVWFEDEFTPWRAHLEQASAL
Mb1779c|M.bovis_AF2122/97 -----------MTDTIQSLLRQHVSDPTIAVKYGGLQWTWSQYLAESAAR
Rv1750c|M.tuberculosis_H37Rv -----------MTDTIQSLLRQHVSDPTIAVKYGGLQWTWSQYLAESAAR
MAV_5257|M.avium_104 ----------MAEDTIQALLRKRWSDPGVAVKYGDTQWSWAQYLRDAAAR
MSMEG_4952|M.smegmatis_MC2_155 -----------MADTLQQLLRSRAEQDTVAVKYGDRTWTWREHIAEAAAQ
MLBr_02661|M.leprae_Br4923 MKSESITSSRAPVEQIDARLTGQFESPSIASLVEAAAIRNPSAPALVVTD
: * . .
Mflv_1548|M.gilvum_PYR-GCK ASLATRLLDGV---------------PGPFHVGILLANVPDFVFWLGGAA
Mvan_5209|M.vanbaalenii_PYR-1 ASIASTLSD-----------------LDPPHIGVLLDNVPDFVFWLGGAA
MAB_3628c|M.abscessus_ATCC_199 GALARKLRAD-----------------GPFHIGVLLDNVPDFVFWLGGAA
MMAR_4994|M.marinum_M AAALRERLDP----------------TRPPHVGVLLQNTPFFSAMLAAAG
MUL_4069|M.ulcerans_Agy99 AAALRERLDP----------------TRPPHVGVLLRNTPFFSAMLTAAG
TH_1138|M.thermoresistible__bu AAALTARLDP----------------NRPPHVGVLSGNTPMFSALLVAAA
Mb1779c|M.bovis_AF2122/97 AAALITIADP----------------QRPTHIGSLLGNTPEMLAQLAAAG
Rv1750c|M.tuberculosis_H37Rv AAALITIADP----------------QRPTHIGSLLGNTPEMLAQLAAAG
MAV_5257|M.avium_104 AAALLGAADP----------------GRPMHVGTLLGNTPEMLSQMAAAG
MSMEG_4952|M.smegmatis_MC2_155 ARFLISRADH----------------GRPLHVGVLLGNTPQMLTALAAAA
MLBr_02661|M.leprae_Br4923 NRIVVSYRDLLRLVDDLTVQLALGGLLPGDRVALCAASNIEFVVGLLAAS
::. . : : .*.
Mflv_1548|M.gilvum_PYR-GCK LAGATVVGLNPTRGTADLAADIRHADCRLIVTDSVGAQRLRG--------
Mvan_5209|M.vanbaalenii_PYR-1 LAGATIVGLNPTRGAADLAADIRHADCRLIVTDAAGADRLRG--------
MAB_3628c|M.abscessus_ATCC_199 LAGATIVGINPTRGPAEMAAEIRHTDCQLIVTDTAHLGRLQS--------
MMAR_4994|M.marinum_M MSGIIPVGLNPVRRGAALARDIEHADCQMVLADTASASTLDG--------
MUL_4069|M.ulcerans_Agy99 MSGIIPVGLNPVRRGAALARDIEHADCQMVLADTASASTLDG--------
TH_1138|M.thermoresistible__bu MTGLVPVGLNPTRRGAALVRDIERADCRLVLADDP-ESVPAG--------
Mb1779c|M.bovis_AF2122/97 LGGYVLCGLNTTRRGDALAADVRRADCQIVVTDADHRALLDG--------
Rv1750c|M.tuberculosis_H37Rv LGGYVLCGLNTTRRGDALAADVRRADCQIVVTDADHRALLDG--------
MAV_5257|M.avium_104 LGGYVLCGLNTTRRGKALAADVRRADCQFVVTDAEHRPLLDG--------
MSMEG_4952|M.smegmatis_MC2_155 LGGYVLCGINTTRRGEGLARDIARVDCQLLITDAEHRGLIDG--------
MLBr_02661|M.leprae_Br4923 RAGLIVVPLDPALPVNEQCIRSQAAGVRVTLVDSLALEGVSDQRAATMRY
* ::.. .. :. :.* .
Mflv_1548|M.gilvum_PYR-GCK --IGHGVPDDRVLVVDDPEYRRALEACATEPAVAAGVGASSLFLLLFTSG
Mvan_5209|M.vanbaalenii_PYR-1 --LDHGVPPDHLLRVDSPDHAQLLDAHRVEPVAAPGVGAKSLFLLLFTSG
MAB_3628c|M.abscessus_ATCC_199 --LELGLSADRLLVIDSPEYLQRIAESRCTAQASEGVGADSLLLLLFTSG
MMAR_4994|M.marinum_M --IEH---VNVDSAEWA----EVIDAHRNSEISFQNAAPADLFMLIYTSG
MUL_4069|M.ulcerans_Agy99 --IEH---VNVDSAEWA----EVIDAHRNSEISFQNAAPADLFMLIYTSG
TH_1138|M.thermoresistible__bu --VDF---IDVTAPRFA----DELAAHRGAPVRFRRAAPDDLFMLIFTSG
Mb1779c|M.bovis_AF2122/97 --LDL---AGARILDTSTPRWAELVAGDGAFVPYREVDTMDPFMMIFTSG
Rv1750c|M.tuberculosis_H37Rv --LDL---AGARILDTSTPRWAELVAGDGAFVPYREVDTMDPFMMIFTSG
MAV_5257|M.avium_104 --LDL---AGTQILDTSTPDWAGFVDAAGELTPHREVAAMDPFMMIFTSG
MSMEG_4952|M.smegmatis_MC2_155 --LEL---PGVTVLDAS-----DICPEAGELTPYREAGPDDTFMMIFTSG
MLBr_02661|M.leprae_Br4923 WPIAVSYGSVTGASEGSLLVHLDDTAALHPVTSTPDGLRHDDAMIMFTGG
: . ::::*.*
Mflv_1548|M.gilvum_PYR-GCK TTGASKAVKCSQGRLARIAYVATEKFGHHRGDVDYCCMPLFHGNALMALW
Mvan_5209|M.vanbaalenii_PYR-1 TTGTSKAVRCSQGRLARIAYAATDKFGHHRDDVDYCCMPLFHGNAIMALW
MAB_3628c|M.abscessus_ATCC_199 TTGASKAVKCSQGRLAQIAHLATEKFGHVRSDVDYCCMPLFHGNALMALW
MMAR_4994|M.marinum_M TSGDPKAVKCSHSKVAIAGVTMTQRFGLGRDDVCYVSMPLFHSNAVLVGW
MUL_4069|M.ulcerans_Agy99 TSGDPKAVKCGHSKVAIAGVTMTQRFGLGRDDVCYVSMPLFHSNAVLVGW
TH_1138|M.thermoresistible__bu TSGDPKAVRVTHDKVAHPGTMLATRFGLGPADTCYLSMPLFHSNAIMAGW
Mb1779c|M.bovis_AF2122/97 TSGNPKAVPVSHLMATFAGRSLTERFGLTEQDTCYVSMPLFHSNAVVAGW
Rv1750c|M.tuberculosis_H37Rv TSGNPKAVPVSHLMATFAGRSLTERFGLTEQDTCYVSMPLFHSNAVVAGW
MAV_5257|M.avium_104 TSGNPKAVQVSHLMAVFAGSNLVQRFGLTEHDTCYVSMPLFHSNAVVAGW
MSMEG_4952|M.smegmatis_MC2_155 TSGDPKAVEVPHAMVLFAGSALVERYGLTCEDVCYLAMPLFHSNAVYAGW
MLBr_02661|M.leprae_Br4923 TTGLPKMVPWTDGNIAGSVHAIITAYQLGPQDATVVVMPLYHGHGLIAAL
*:* .* * . : *. ***:*.:.: .
Mflv_1548|M.gilvum_PYR-GCK APALANG--ATVCLTPSFSASRFLSDIRYFGATFFTYVGKALAYLMATPE
Mvan_5209|M.vanbaalenii_PYR-1 APALANG--ATVCLTPTFSASRFLPDVRYFGATFFTYVGKALGYLMATPE
MAB_3628c|M.abscessus_ATCC_199 APALANG--ATVCLTPTFSASRFLPDVRYFGATFFTYVGKALGYLLATPE
MMAR_4994|M.marinum_M AVAVACQ--GSMALRRKFSASQFLSDVRRYGATYANYVGKPLSYVLATPE
MUL_4069|M.ulcerans_Agy99 AVAVACQ--GSMALRRKFSASQFLSDVRRYGATYANYVGKPLSYVLATPE
TH_1138|M.thermoresistible__bu AVAVAAG--ASVALRRRFSASQFIPDVRRYRASYANYVGKPLSYILATPP
Mb1779c|M.bovis_AF2122/97 APAVVSG--AAIAP-ATFSATGFLDDVRRYHATYMNYVGKPLAYILATPE
Rv1750c|M.tuberculosis_H37Rv APAVVSG--AAIAP-ATFSATGFLDDVRRYHATYMNYVGKPLAYILATPE
MAV_5257|M.avium_104 APAVCSG--AAIVP-AKFSARNFLDDIRRYGATYMNYVGKPLAYVLATPE
MSMEG_4952|M.smegmatis_MC2_155 SVAVGAG--AAMAP-ATFSASGFLPDIRRYGATYMNYVGKPLAYILATPE
MLBr_02661|M.leprae_Br4923 LSTLASGGVVLLPARARFSARTFWDDIDAVAATWYTAAPAIHRILLELAS
:: : *** * *: *:: . . :: .
Mflv_1548|M.gilvum_PYR-GCK ----QADDSDNTLTRGFGTEASPEDQAEFRRRFGAELHEGYGSSEGGAVA
Mvan_5209|M.vanbaalenii_PYR-1 ----APDDADNTLQRGFGTEASPEDQKEFRRRFAAELFEGYGSSEGGAVA
MAB_3628c|M.abscessus_ATCC_199 ----RADDADNQLARGFGTEASPEDQAQFRRRFGAELFEGYGSSEGGGAV
MMAR_4994|M.marinum_M ----RPDDADNPLRAVYGNEGVPGDSERFGCRFGCVVQDGFGSTEGGVAI
MUL_4069|M.ulcerans_Agy99 ----RPDDADNPLRAVYGNEGVPGDIERFGCRFGCVVQDGFGSTEGGVAI
TH_1138|M.thermoresistible__bu ----RADDADNPLRILYGNEGAPRDIARFARRFDVLVVDGFGSTEGGVAI
Mb1779c|M.bovis_AF2122/97 ----RDDDADNPLRVAFGNEANDKDIEEFSRRFGVQVEDGFGSTENAVIV
Rv1750c|M.tuberculosis_H37Rv ----RDDDADNPLRVAFGNEANDKDIEEFSRRFGVQVEDGFGSTENAVIV
MAV_5257|M.avium_104 ----RADDADNPLRVAFGNEANDKDIEEFGRRFGVQVEDGFGSTENAVIV
MSMEG_4952|M.smegmatis_MC2_155 ----KPDDHDNPLRIAFGNEAADRDIAAFSRRFGCTVWDGFGSTETAVII
MLBr_02661|M.leprae_Br4923 TQSFRSKRAKLRFIRSCSAPLTQETAQALREEFLAPVICAFGMTEATHQV
. . : . : .* : .:* :*
Mflv_1548|M.gilvum_PYR-GCK TPDPAAPATALGRPAHEDVVVVDPATLKPCAPAVLDPRGRVTNADDAIGE
Mvan_5209|M.vanbaalenii_PYR-1 TPDPAAPPAALGRPAHDDVVVVHPETLCLCPTAVLDASGRVLNPDEAVGE
MAB_3628c|M.abscessus_ATCC_199 VLDPAAPPGALGRPAHEGVVVVDPETLQDCAAAVFDQHGRVLNADAAVGE
MMAR_4994|M.marinum_M ARTPDTPAGSLG-PLPENIEILDPETGQQCPVG-------------AVGE
MUL_4069|M.ulcerans_Agy99 ARTPDTPAGSLG-PLPENIEIVDPETGQQCPVG-------------AVGE
TH_1138|M.thermoresistible__bu ARTPDTPDGALG-PLTDEVAIIDVDTGEPCPPG-------------KIGE
Mb1779c|M.bovis_AF2122/97 IREPGTPPGSIG-RGAHGVAVYNGETVTECAVARFDAHGALTNADEAIGE
Rv1750c|M.tuberculosis_H37Rv IREPGTPPGSIG-RGAHGVAVYNGETVTECAVARFDAHGALTNADEAIGE
MAV_5257|M.avium_104 IREPGTPRGSIG-RGIDGVAIYNSDTVTECPVARFDADGALINADEAVGE
MSMEG_4952|M.smegmatis_MC2_155 TRPDDCPPGSIG-KGFPGVAIYDPQTVTECAVAEFDATGALANPDAAIGE
MLBr_02661|M.leprae_Br4923 TTTNIKWFGQGENPTVTNGLVGQSTGVQIRIVG----SDGQPLPPDTVGE
: . . :**
Mflv_1548|M.gilvum_PYR-GCK IVDRRGARDFEGYYRNDAADADRVRNGWYWSGDLGYVDRDGFLYFAGRRG
Mvan_5209|M.vanbaalenii_PYR-1 IVDRRGAADFEGYYRNDAADAERVRNGWYWSGDLGYLDGAGFLYFAGRRG
MAB_3628c|M.abscessus_ATCC_199 IVDKQGRRGFEGYYNNDDADADRIRNGWYWTGDLGYLDAAGFIYFAGRRG
MMAR_4994|M.marinum_M LVNTAGPGRFEGYYNDEAASAQRMSGGVYHSGDLAYRDAAGYAYFAGRLG
MUL_4069|M.ulcerans_Agy99 LVNTAGPGRFEGYYNDEAASAQRMSGGVYHSGDLAYRDAAGYAYFAGRLG
TH_1138|M.thermoresistible__bu IVNPTGPGWFRGYYNDPDAEAERMAGGVYHTGDLGYCDENGYIYFAGRLG
Mb1779c|M.bovis_AF2122/97 LVNTTGSGFFTGYYNDPEANAERMRHGMYWSGDLAYRDSEGWIYLAGRTA
Rv1750c|M.tuberculosis_H37Rv LVNTTGSGFFTGYYNDPEANAERMRHGMYWSGDLAYRDSEGWIYLAGRTA
MAV_5257|M.avium_104 LVNTAGSGFFTGYYNDPDANAERMRHGMYWSGDLAYRDADGWIYLAGRTA
MSMEG_4952|M.smegmatis_MC2_155 LVNTAGGGLFRGYYNDQGATDQRMRHGMYWSGDLAYRDADGWIYLAGRTA
MLBr_02661|M.leprae_Br4923 VWLR-GSTVVRGYLGDPAITAANFTHGWLRTGDLGSLSVTGDLRIRGRIK
: * . ** : .. * :***. . * : **
Mflv_1548|M.gilvum_PYR-GCK DWIRVDGENTSALTIERVLRRHPEVIAAAVYGVPDPRSGDQVMAAVEVAE
Mvan_5209|M.vanbaalenii_PYR-1 DWLRVDGENISTLTIERVLRRHAGVIAAAVYGVPDPRSGDQMMAAVEVAD
MAB_3628c|M.abscessus_ATCC_199 DWIRVDGENMSALTVERVLRRHPLVIAAGVYAVPDPRSGDQVMASIEVAE
MMAR_4994|M.marinum_M DWMRVDGENLGTAPIERVLLRYPDAIEVAVYAIPDPVVGDQVMAAMVLTP
MUL_4069|M.ulcerans_Agy99 DWMRVDGENLGTAPIERVLLRYPDAIEVAVYAIPDPVVGDQVMAAMVLTP
TH_1138|M.thermoresistible__bu DWMRVDGENLGTAPIERILLRHPDVVEAAVYPIPDPAVGDRVMAALVLTD
Mb1779c|M.bovis_AF2122/97 DWMRVDGENLTAAPIERILLRYKAINRVAVYAVPDEYVGDQVMAALVLRA
Rv1750c|M.tuberculosis_H37Rv DWMRVDGENLTAAPIERILLRYKAINRVAVYAVPDEYVGDQVMAALVLRA
MAV_5257|M.avium_104 DWMRVDGENLAAAPIERILLRHTAINRVAVYAVPDGHVGDQVMAAVVLND
MSMEG_4952|M.smegmatis_MC2_155 DWMRVDGENLTAAPIERILLRLDAVNRVAVYPVPDEYVGDQVMAAIVLQD
MLBr_02661|M.leprae_Br4923 ELINRSGEKISPERVEGVLASHHNVMEVAVFGDPDKVYGETVTAVIVPRE
: :. .**: . :* :* ..*: ** *: : * :
Mflv_1548|M.gilvum_PYR-GCK PGSFDINAFVDFLVGQDDLGAKGFPRLLRVSARLPATGSNKVLKRELQTQ
Mvan_5209|M.vanbaalenii_PYR-1 PGSFDVDAFASFLVAQDDLGRKGFPRLLRISAKLPATGSNKVLKRDLQAQ
MAB_3628c|M.abscessus_ATCC_199 PDSFDVDGFTQYLSAQEELGSKGIPRFLRISANLPVTGSNKVLKRELQEQ
MMAR_4994|M.marinum_M GAKFDVDKFRAFLAGQSDLGPKQWPSYVRICSALPRTETFKVLKRQLAAD
MUL_4069|M.ulcerans_Agy99 GAKFDVDKFRAFLAGQSDLGPKQWPSYVRICSALPRTETFKVLKRQLAAD
TH_1138|M.thermoresistible__bu GAEFDPGRFRDFLADQPDLGPKQWPSFVRVAEVLPRTETFKVLKRQLSAE
Mb1779c|M.bovis_AF2122/97 GDTFDPDAFEAFLDAQPDLSTKARPRYIRIAADLPSTATHKVLKRQLIDE
Rv1750c|M.tuberculosis_H37Rv GDTFDPDAFEAFLDAQPDLSTKARPRYIRIAADLPSTATHKVLKRQLIDE
MAV_5257|M.avium_104 GETLTPAAFEVFLDAQPDLSPKARPRYVRIAADLPSTATHKVLKRQLISQ
MSMEG_4952|M.smegmatis_MC2_155 GATLTPGEFEEFLAAQRDLSPKAWPRYVWIAEDLPTTATNKILKRELVAT
MLBr_02661|M.leprae_Br4923 VIAPTPSELAVFCRDR--LAAFEVPTRFQEASALPHTAKGSLDRRAVAEQ
: : : *. * . . ** * . .: :* :
Mflv_1548|M.gilvum_PYR-GCK RWHTDEAVYRWSGRGRPRYRLMTADDRAALDEEFVTHGRAHHGTT
Mvan_5209|M.vanbaalenii_PYR-1 RWHTDDAVHRWVGRGHPRYRLMTDADRGALDAEFTVHGREHHVAE
MAB_3628c|M.abscessus_ATCC_199 RWHTDEPVFRWVGRGGPVFLRMSSDDKTALDAQFAVYGRQRLL--
MMAR_4994|M.marinum_M GVDC--GDPVWPIR-----DQS-----------------------
MUL_4069|M.ulcerans_Agy99 GVDC--GDPVWPIR-----DQS-----------------------
TH_1138|M.thermoresistible__bu ATDC--ADPVWPIER-PRGDRCASP--------------------
Mb1779c|M.bovis_AF2122/97 GTAVGKADTLWVRE--PRGSAYHHASGPAKAI-------------
Rv1750c|M.tuberculosis_H37Rv GTAVGKADTLWVRE--PRGSAYHHASGPAKAI-------------
MAV_5257|M.avium_104 GTAVAAGEVLWERE--PRGTAYTVVASPDCGVPGDGSAPSTVAPV
MSMEG_4952|M.smegmatis_MC2_155 GTDP-AGRVLWHRDG-TRFTDLSAASAPGAAPGISPA--------
MLBr_02661|M.leprae_Br4923 FAHRG----------------------------------------