For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAEMLAGRYELRGLLGRGGMAEVRDGWDTRLDRSVAVKLLYPSQSSDDSVRRRFEDEARSAARLSHPNI VAIHDFGEHDSAPFIVMERLPGRTLADVIEQGPMPAPQVRRVLDEVLAALSVAHAAGVLHRDIKPANILL TTAGDSVKVADFGIAKSAGAAHTATGQIVGTLCYLSPERVMGAPASVADDLYAVGVMGYEALLGRRAFPQ DNPAALARAIIDVPPPPLRALSVTADPALVAVIDRAMSRDPRLRFGSAPQMRAALAGDPLALNNAPPPAP PPRAGTRVLAQPLPPSAGFTSAPVGYPTAAPPPPTRARMYLMAVAAVLAFTVAALALALDPFSSPPPMQT VGTSTTAPPPPTSVPPPTSAAPLPPPIVPVTQAPPPDNGPKKGPDKKGEKGNNGRGNGNKPD
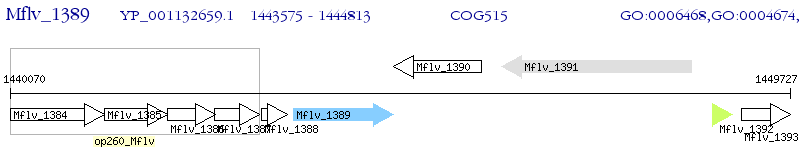
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1389 | - | - | 100% (412) | protein kinase |
| M. gilvum PYR-GCK | Mflv_1671 | - | 2e-83 | 56.89% (283) | protein kinase |
| M. gilvum PYR-GCK | Mflv_0811 | - | 2e-54 | 35.24% (403) | protein kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0015c | pknA | 2e-55 | 36.67% (390) | transmembrane serine/threonine-protein kinase A PKNA |
| M. tuberculosis H37Rv | Rv0015c | pknA | 2e-55 | 36.67% (390) | transmembrane serine/threonine-protein kinase A PKNA |
| M. leprae Br4923 | MLBr_00017 | pknA | 2e-52 | 36.01% (386) | putative serine/threonine protein kinase |
| M. abscessus ATCC 19977 | MAB_0034c | - | 3e-55 | 36.10% (421) | putative serine/threonine-protein kinase PknA |
| M. marinum M | MMAR_0017 | pknA | 2e-55 | 37.85% (391) | serine/threonine-protein kinase a PknA |
| M. avium 104 | MAV_0019 | - | 6e-57 | 36.20% (395) | serine/threonine protein kinases Drp72 |
| M. smegmatis MC2 155 | MSMEG_6206 | - | 1e-103 | 50.12% (411) | serine/threonine protein kinase PksC |
| M. thermoresistible (build 8) | TH_2610 | - | 2e-51 | 41.24% (291) | PUTATIVE serine/threonine protein kinase |
| M. ulcerans Agy99 | MUL_0019 | pknA | 2e-55 | 38.01% (392) | serine/threonine-protein kinase a PknA |
| M. vanbaalenii PYR-1 | Mvan_5401 | - | 1e-145 | 66.83% (416) | protein kinase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0017|M.marinum_M ------MSPRVGMTLSGRYRLQRLIATGGMGQVWEAVDSRLGRRVAVKVL
MUL_0019|M.ulcerans_Agy99 ------MSPIVGMTLSGRYRLQRLIATGGMGQVWEAVDSRLGRRVAVKVL
MAV_0019|M.avium_104 ------------MTLSGRYRLQRLIATGGMGQVWEAVDNRLGRRVAVKVL
Mb0015c|M.bovis_AF2122/97 ------MSPRVGVTLSGRYRLQRLIATGGMGQVWEAVDNRLGRRVAVKVL
Rv0015c|M.tuberculosis_H37Rv ------MSPRVGVTLSGRYRLQRLIATGGMGQVWEAVDNRLGRRVAVKVL
MLBr_00017|M.leprae_Br4923 ------MSPRIGVMLSGRYRLHRLIATGGMGQVWEAVDSRLGRRVAVKVL
MAB_0034c|M.abscessus_ATCC_199 ------MTVRVGETLSGRYRLQRLIATGGMGQVWEGTDSRLGRKVAVKVL
TH_2610|M.thermoresistible__bu VETYQQSDPFAGTVLDGRYRIDTLIATGGMSAVYRGLDLRLDRPVAVKVM
Mflv_1389|M.gilvum_PYR-GCK --------MT-AEMLAGRYELRGLLGRGGMAEVRDGWDTRLDRSVAVKLL
Mvan_5401|M.vanbaalenii_PYR-1 --------MTGRETLVGRYELRGLLGCGGMAEVRDGWDIRLDRPVAVKLL
MSMEG_6206|M.smegmatis_MC2_155 ----MGSAERFQRLLGDRYELRGVLGRGGMAEVRDGWDTRLARPVAIKLL
* .**.: ::. ***. * . * ** * **:*::
MMAR_0017|M.marinum_M KQEFSSDPEFIERFRAEARTTAMLNHPGIAAVHDYGESQMNGEGRTAYLV
MUL_0019|M.ulcerans_Agy99 KQEFSSDPEFIERFRAEARTTAMLNHPGIAAVHDYGESQMNGEGRTAYLV
MAV_0019|M.avium_104 KQEFSQDPEFIERFRAEARTTAMLNHPGIAAVHDYGESQLDGEGRTAYLV
Mb0015c|M.bovis_AF2122/97 KSEFSSDPEFIERFRAEARTTAMLNHPGIASVHDYGESQMNGEGRTAYLV
Rv0015c|M.tuberculosis_H37Rv KSEFSSDPEFIERFRAEARTTAMLNHPGIASVHDYGESQMNGEGRTAYLV
MLBr_00017|M.leprae_Br4923 KGEFSSDPEFIERFRAEARTTAMLNHPGIASVHDYGESHMDGEGRTAYLV
MAB_0034c|M.abscessus_ATCC_199 KAEFSSDPEFIERFRAEARTVAMLNHPGIASVYDYGETDIDSEGRTAYLV
TH_2610|M.thermoresistible__bu DSRYAGDQQFLTRFQREARAAARLKDPGLVAVYDQGPG--SPASHPPYLV
Mflv_1389|M.gilvum_PYR-GCK YPSQSSDDSVRRRFEDEARSAARLSHPNIVAIHDFGEHD-----SAPFIV
Mvan_5401|M.vanbaalenii_PYR-1 HPNFNADADARARFCDEARSAAALSHPNIVAVHDFGEHD-----GRPFIV
MSMEG_6206|M.smegmatis_MC2_155 YPALAADADLRARFEREARAAAALTHPHVVAVHDCGEHA-----GTPYIV
* . ** ***:.* *..* :.:::* * .::*
MMAR_0017|M.marinum_M MELVNGEPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRDVKP
MUL_0019|M.ulcerans_Agy99 MELVNGEPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRDVKP
MAV_0019|M.avium_104 MELVNGEPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRDVKP
Mb0015c|M.bovis_AF2122/97 MELVNGEPLNSVLKRTGRLSLRHALDMLEQTGRALQIAHAAGLVHRDVKP
Rv0015c|M.tuberculosis_H37Rv MELVNGEPLNSVLKRTGRLSLRHALDMLEQTGRALQIAHAAGLVHRDVKP
MLBr_00017|M.leprae_Br4923 MELVNGEPLNSVLKRTGRLSLRHALDMLEQTGRALQIAHAAGLVHRDVKP
MAB_0034c|M.abscessus_ATCC_199 MELIHGEPLNSVLKRTGRLSLRHSLDMLEQTGRALQVAHAAGLVHRDVKP
TH_2610|M.thermoresistible__bu MELIEGGTLRELLRERGPMPPHAAAAVLAPVLGGLAVAHRAGLVHRDIKP
Mflv_1389|M.gilvum_PYR-GCK MERLPGRTLADVIEQ-GPMPAPQVRRVLDEVLAALSVAHAAGVLHRDIKP
Mvan_5401|M.vanbaalenii_PYR-1 MERLPGRTLADLIAQ-GPMAPAQVRAMLDDVLAALDVAHTAGVVHRDIKP
MSMEG_6206|M.smegmatis_MC2_155 MERLSGATLADEIAA-GPLPQERVAELLDQILSALIAAHAAGIVHRDIKP
** : * .* . : * :. :* .* ** **::***:**
MMAR_0017|M.marinum_M GNILITPTG-QVKITDFGIAKAVDAAPVTQTGMVMGTAQYIAPEQALGQD
MUL_0019|M.ulcerans_Agy99 GNILITPTG-QVKITDFGIAKAVDAAPVTQTGMVMGTAQYIAPEQALGQD
MAV_0019|M.avium_104 GNILITPTG-QVKITDFGIAKAVDAAPVTQTGMVMGTAQYIAPEQALGHD
Mb0015c|M.bovis_AF2122/97 GNILITPTG-QVKITDFGIAKAVDAAPVTQTGMVMGTAQYIAPEQALGHD
Rv0015c|M.tuberculosis_H37Rv GNILITPTG-QVKITDFGIAKAVDAAPVTQTGMVMGTAQYIAPEQALGHD
MLBr_00017|M.leprae_Br4923 GNILITPTG-QVKITDFGIAKAVDAAPVTQTGMVMGTAQYIAPEQALGHD
MAB_0034c|M.abscessus_ATCC_199 GNILITPTG-QVKLTDFGIAKAVDAAPVTQTGMVMGTAQYIAPEQALGHD
TH_2610|M.thermoresistible__bu ENVLISDDG-EVKIADFGLVRAVAEAKITSDSVILGTAAYLSPEQVATGD
Mflv_1389|M.gilvum_PYR-GCK ANILLTTAGDSVKVADFGIAKSA-GAAHTATGQIVGTLCYLSPERVMGAP
Mvan_5401|M.vanbaalenii_PYR-1 GNILLSATGDRMQVADFGIAKSG-GAAHTMTGQIVGTLCYMSPERVSGAP
MSMEG_6206|M.smegmatis_MC2_155 GNILIGHTG-QAKLADFGIAKTD-GVPHTQAGQILGTMAYLSPQRVTGQP
*:*: * :::***:.:: . * . ::** *::*::.
MMAR_0017|M.marinum_M ASPSSDVYSLGVVGYEAVSGKRPFTGDGALTVAMKHIK---EPPPPLPAE
MUL_0019|M.ulcerans_Agy99 ASPSSDVYSLGVVGYEAVSGKRPFTGDGALTVAMKHIK---EPPPPLPAE
MAV_0019|M.avium_104 ATPASDVYSLGVVGYEVVSGKRPFSGDGALTVAMKHIK---EPPPPLPAE
Mb0015c|M.bovis_AF2122/97 ASPASDVYSLGVVGYEAVSGKRPFAGDGALTVAMKHIK---EPPPPLPPD
Rv0015c|M.tuberculosis_H37Rv ASPASDVYSLGVVGYEAVSGKRPFAGDGALTVAMKHIK---EPPPPLPPD
MLBr_00017|M.leprae_Br4923 ATPASDVYSLGVIGYEVVSGKRPFTGDGALTVAMKHIK---EPPPPLPAD
MAB_0034c|M.abscessus_ATCC_199 ATPASDVYSLGVVGYEAVSGKRPFSGDGALTVAMKHIK---ETPSPLPAD
TH_2610|M.thermoresistible__bu ATPRSDVYAVGILTYEMLTGNTPFTGDNALALAYQRLDNDVPPPSSAISG
Mflv_1389|M.gilvum_PYR-GCK ASVADDLYAVGVMGYEALLGRRAFPQDNPAALARAIIDVPPPPLRALSVT
Mvan_5401|M.vanbaalenii_PYR-1 ASVADDLYAVGVMGYEALLARRAFPQESPVALVRAIVDAPPPALAGLTGG
MSMEG_6206|M.smegmatis_MC2_155 AGFADDLYAVGVVGYEALTGRRPFDRDNPAAMVRAILDDVPVPIASVRSD
* .*:*::*:: ** : .. .* :.. ::. :. .
MMAR_0017|M.marinum_M LPPNVRELIEITLVKNPGLRYRSGGPFADAVAAVRAGRRPPRPSQSP--P
MUL_0019|M.ulcerans_Agy99 LPPNVRELIEITLVKNPGLRYRSGGPFADAVAAVRAGRRPPRPSQSP--P
MAV_0019|M.avium_104 LPPNVRELIEITLVKNPAMRYRSGGPFADAVAAVRAGRRPPRPSQSP--P
Mb0015c|M.bovis_AF2122/97 LPPNVRELIEITLVKNPAMRYRSGGPFADAVAAVRAGRRPPRPSQTP--P
Rv0015c|M.tuberculosis_H37Rv LPPNVRELIEITLVKNPAMRYRSGGPFADAVAAVRAGRRPPRPSQTP--P
MLBr_00017|M.leprae_Br4923 LPPNVRELIEITLVKNPGMRYPSGGLFAEAVAAVRAGHRPPRPNQTP--S
MAB_0034c|M.abscessus_ATCC_199 LPPNVRELIEITLVKNPQNRYTSGGPFADAVAAVRAGRRPPRPNQAP--T
TH_2610|M.thermoresistible__bu VPPQFDELVATATARDPADRFADAHDMAAAVADLVAELELP-PFRVP--A
Mflv_1389|M.gilvum_PYR-GCK ADPALVAVIDRAMSRDPRLRFGSAPQMRAALAGDPLALNNAPPPAPPPRA
Mvan_5401|M.vanbaalenii_PYR-1 ADPVLAAVIDRAMSRDPRRRFGSAAQMRAALAGDVTALSTAAEPRP----
MSMEG_6206|M.smegmatis_MC2_155 VAPGLIHVIDMAMAREVAFRYPDAATMRAALTGAEPPTQRVG-RRQ----
* . :: : :: *: .. : *::
MMAR_0017|M.marinum_M AGRAAPAAIPSSAPARIAPAPTTRTTTPR-RTRPATGGHRPPPA-RRTFS
MUL_0019|M.ulcerans_Agy99 AGRAAPAAIPSSAPARIAPAPTTRTTTPR-RTRPATGGHRPPPS-RRTFS
MAV_0019|M.avium_104 PGRASPAAIPSSPTTRAAAVSSGRTAAPR-RTRPATGGHRPPPA-RRTFS
Mb0015c|M.bovis_AF2122/97 PGRAAPAAIPSGTTARVAANSAGRTAASR-RSRPATGGHRPP---RRTFS
Rv0015c|M.tuberculosis_H37Rv PGRAAPAAIPSGTTARVAANSAGRTAASR-RSRPATGGHRPP---RRTFS
MLBr_00017|M.leprae_Br4923 SGRASPTTIPSSTQARAAVACGTKTPAPR-RSRPSTSGNRPPPA-RNTFS
MAB_0034c|M.abscessus_ATCC_199 ISRATPAAIPPATQVRPAVGAPSRVAPPTSRTRQPTGSHRPPPPRRGTFS
TH_2610|M.thermoresistible__bu PR--------NSAQHLSAIRQHSQSARPT-------TTLQPPX-------
Mflv_1389|M.gilvum_PYR-GCK GTRVLAQPLPPSAGFTSAPVGYPTAAPP-----PPTRARMYLMAVAAVL-
Mvan_5401|M.vanbaalenii_PYR-1 LTKVLAAPLPPSA-------AYPAPAPRR----PATRARRYVVGAAAVMT
MSMEG_6206|M.smegmatis_MC2_155 -TKVLAAPLPPIP---------PPPVLPS----PRHWRTRLILAVLGIV-
.
MMAR_0017|M.marinum_M SGQRALLWAAGVLGALAIIIAVLIVINSRAD-SQQQPPPPTVTQTTTAHQ
MUL_0019|M.ulcerans_Agy99 SGQRALLWAAGVLGALAIIIAVLIVINSRAD-SQQQSPPPTVTQTTTAHQ
MAV_0019|M.avium_104 SGQRALLWAAGVLGALAIIIAVLIVINSRAD-QQQQ--PPTVTDTG----
Mb0015c|M.bovis_AF2122/97 SGQRALLWAAGVLGALAIIIAVLLVIKAPGDNSPQQAPTPTVTTTG----
Rv0015c|M.tuberculosis_H37Rv SGQRALLWAAGVLGALAIIIAVLLVIKAPGDNSPQQAPTPTVTTTG----
MLBr_00017|M.leprae_Br4923 SGQRALLWAAGMLGALAIIIAVLIVINSYAGNEQHQPPTPTVTDTG----
MAB_0034c|M.abscessus_ATCC_199 PGQKALMWAAAVLGIFAIIIAALILLQASEKKNQTPSIPATVSGSE----
TH_2610|M.thermoresistible__bu ------XXGAGGRARVATEIGDRIGIVDQPPTARGRHANAVRTAFR----
Mflv_1389|M.gilvum_PYR-GCK AFTVAALALALDPFSSPPP-MQTVGTSTTAPPPPTSVPPPTSAAPLPPPI
Mvan_5401|M.vanbaalenii_PYR-1 AFVISALALAMDPFSSSAPRPEPVSTSTSAPSPTTPPPPPITTPPPSP--
MSMEG_6206|M.smegmatis_MC2_155 STLILVLVLALARGSSGLTSPTSTPTATTVPPATSAVVPPPVPSTPVPAA
* . .
MMAR_0017|M.marinum_M TPTGQGPRLNWTEGKAIGNPGLQSNQPDLVDDAGRNRPIGNR-GSATSWN
MUL_0019|M.ulcerans_Agy99 TPTGQGPRLNWTDGQSIGNPGLQSNQPDLVDDAGRNRPIGNR-DSATSWN
MAV_0019|M.avium_104 TPPASAPPPTKTPSGSGAHPGLRLDWP----DDGTIGASGFRDGPARHWT
Mb0015c|M.bovis_AF2122/97 NPPASN------TGGTDASP--RLNWT----ERGETRHSGLQ-----SWV
Rv0015c|M.tuberculosis_H37Rv NPPASN------TGGTDASP--RLNWT----ERGETRHSGLQ-----SWV
MLBr_00017|M.leprae_Br4923 TPPATKT----LSGFPAAYCEYRVNWT----NHKEISNSGLP-----KQA
MAB_0034c|M.abscessus_ATCC_199 TPSSPGS----ASPGALAPANIVALLP----DAEPDPIPGTK--------
TH_2610|M.thermoresistible__bu -------------------------------NDE----------------
Mflv_1389|M.gilvum_PYR-GCK VPVTQAPP---PDNGPKKGPDKKGEKGN---NGRGNGNKPD---------
Mvan_5401|M.vanbaalenii_PYR-1 APAVTAAT---QQPEPQQGRDDKGPRGNGRGNGKGKGEKKD---------
MSMEG_6206|M.smegmatis_MC2_155 VPEEPRGPGRGPGPGPGPGKGKGHEKGHGPKGPKGPPHP-----------
MMAR_0017|M.marinum_M VTVTPQHLAALARFEMQR
MUL_0019|M.ulcerans_Agy99 VTVTPQHRAALARFEMQR
MAV_0019|M.avium_104 SQ----------------
Mb0015c|M.bovis_AF2122/97 VPPTPHSRASLARYEIAQ
Rv0015c|M.tuberculosis_H37Rv VPPTPHSRASLARYEIAQ
MLBr_00017|M.leprae_Br4923 ARAQLAGAIDISPVAGQT
MAB_0034c|M.abscessus_ATCC_199 ---SP-------------
TH_2610|M.thermoresistible__bu ------------------
Mflv_1389|M.gilvum_PYR-GCK ------------------
Mvan_5401|M.vanbaalenii_PYR-1 ------------------
MSMEG_6206|M.smegmatis_MC2_155 ------------------