For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GSAERFQRLLGDRYELRGVLGRGGMAEVRDGWDTRLARPVAIKLLYPALAADADLRARFEREARAAAALT HPHVVAVHDCGEHAGTPYIVMERLSGATLADEIAAGPLPQERVAELLDQILSALIAAHAAGIVHRDIKPG NILIGHTGQAKLADFGIAKTDGVPHTQAGQILGTMAYLSPQRVTGQPAGFADDLYAVGVVGYEALTGRRP FDRDNPAAMVRAILDDVPVPIASVRSDVAPGLIHVIDMAMAREVAFRYPDAATMRAALTGAEPPTQRVGR RQTKVLAAPLPPIPPPPVLPSPRHWRTRLILAVLGIVSTLILVLVLALARGSSGLTSPTSTPTATTVPPA TSAVVPPPVPSTPVPAAVPEEPRGPGRGPGPGPGPGKGKGHEKGHGPKGPKGPPHP*
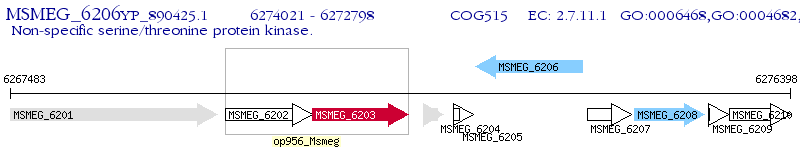
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6206 | - | - | 100% (407) | serine/threonine protein kinase PksC |
| M. smegmatis MC2 155 | MSMEG_0028 | - | 5e-53 | 38.48% (343) | serine-threonine protein kinase |
| M. smegmatis MC2 155 | MSMEG_0030 | pknA | 3e-52 | 32.33% (399) | serine/threonine protein kinase PknA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0015c | pknA | 2e-53 | 33.42% (386) | transmembrane serine/threonine-protein kinase A PKNA |
| M. gilvum PYR-GCK | Mflv_1389 | - | 1e-103 | 50.12% (411) | protein kinase |
| M. tuberculosis H37Rv | Rv0015c | pknA | 3e-53 | 33.42% (386) | transmembrane serine/threonine-protein kinase A PKNA |
| M. leprae Br4923 | MLBr_00017 | pknA | 2e-53 | 34.46% (386) | putative serine/threonine protein kinase |
| M. abscessus ATCC 19977 | MAB_0034c | - | 3e-54 | 33.57% (420) | putative serine/threonine-protein kinase PknA |
| M. marinum M | MMAR_0017 | pknA | 3e-54 | 34.38% (381) | serine/threonine-protein kinase a PknA |
| M. avium 104 | MAV_0019 | - | 5e-55 | 34.15% (410) | serine/threonine protein kinases Drp72 |
| M. thermoresistible (build 8) | TH_2631 | - | 4e-52 | 38.48% (343) | PUTATIVE serine/threonine protein kinase |
| M. ulcerans Agy99 | MUL_0019 | pknA | 6e-55 | 34.65% (381) | serine/threonine-protein kinase a PknA |
| M. vanbaalenii PYR-1 | Mvan_5401 | - | 1e-102 | 53.65% (397) | protein kinase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0017|M.marinum_M --MSPRVGMTLSGRYRLQRLIATGGMGQVWEAVDSRLGRRVAVKVLKQEF
MUL_0019|M.ulcerans_Agy99 --MSPIVGMTLSGRYRLQRLIATGGMGQVWEAVDSRLGRRVAVKVLKQEF
MAV_0019|M.avium_104 --------MTLSGRYRLQRLIATGGMGQVWEAVDNRLGRRVAVKVLKQEF
Mb0015c|M.bovis_AF2122/97 --MSPRVGVTLSGRYRLQRLIATGGMGQVWEAVDNRLGRRVAVKVLKSEF
Rv0015c|M.tuberculosis_H37Rv --MSPRVGVTLSGRYRLQRLIATGGMGQVWEAVDNRLGRRVAVKVLKSEF
MLBr_00017|M.leprae_Br4923 --MSPRIGVMLSGRYRLHRLIATGGMGQVWEAVDSRLGRRVAVKVLKGEF
MAB_0034c|M.abscessus_ATCC_199 --MTVRVGETLSGRYRLQRLIATGGMGQVWEGTDSRLGRKVAVKVLKAEF
Mflv_1389|M.gilvum_PYR-GCK ----MT-AEMLAGRYELRGLLGRGGMAEVRDGWDTRLDRSVAVKLLYPSQ
Mvan_5401|M.vanbaalenii_PYR-1 ----MTGRETLVGRYELRGLLGCGGMAEVRDGWDIRLDRPVAVKLLHPNF
MSMEG_6206|M.smegmatis_MC2_155 MGSAERFQRLLGDRYELRGVLGRGGMAEVRDGWDTRLARPVAIKLLYPAL
TH_2631|M.thermoresistible__bu ----MTTPQHLSDRYELGEILGFGGMSEVHLARDLRLHRDVAVKVLRADL
* .**.* ::. ***.:* . * ** * **:*:*
MMAR_0017|M.marinum_M SSDPEFIERFRAEARTTAMLNHPGIAAVHDYGESQMNGEGRTAYLVMELV
MUL_0019|M.ulcerans_Agy99 SSDPEFIERFRAEARTTAMLNHPGIAAVHDYGESQMNGEGRTAYLVMELV
MAV_0019|M.avium_104 SQDPEFIERFRAEARTTAMLNHPGIAAVHDYGESQLDGEGRTAYLVMELV
Mb0015c|M.bovis_AF2122/97 SSDPEFIERFRAEARTTAMLNHPGIASVHDYGESQMNGEGRTAYLVMELV
Rv0015c|M.tuberculosis_H37Rv SSDPEFIERFRAEARTTAMLNHPGIASVHDYGESQMNGEGRTAYLVMELV
MLBr_00017|M.leprae_Br4923 SSDPEFIERFRAEARTTAMLNHPGIASVHDYGESHMDGEGRTAYLVMELV
MAB_0034c|M.abscessus_ATCC_199 SSDPEFIERFRAEARTVAMLNHPGIASVYDYGETDIDSEGRTAYLVMELI
Mflv_1389|M.gilvum_PYR-GCK SSDDSVRRRFEDEARSAARLSHPNIVAIHDFGEHDS-----APFIVMERL
Mvan_5401|M.vanbaalenii_PYR-1 NADADARARFCDEARSAAALSHPNIVAVHDFGEHDG-----RPFIVMERL
MSMEG_6206|M.smegmatis_MC2_155 AADADLRARFEREARAAAALTHPHVVAVHDCGEHAG-----TPYIVMERL
TH_2631|M.thermoresistible__bu ARDPSFYLRFRREAQNAAALNHPAIVAVYDTGEAETP-TGPLPYIVMEYV
* . ** **: .* *.** :.:::* ** .::*** :
MMAR_0017|M.marinum_M NGEPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRDVKPGNIL
MUL_0019|M.ulcerans_Agy99 NGEPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRDVKPGNIL
MAV_0019|M.avium_104 NGEPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRDVKPGNIL
Mb0015c|M.bovis_AF2122/97 NGEPLNSVLKRTGRLSLRHALDMLEQTGRALQIAHAAGLVHRDVKPGNIL
Rv0015c|M.tuberculosis_H37Rv NGEPLNSVLKRTGRLSLRHALDMLEQTGRALQIAHAAGLVHRDVKPGNIL
MLBr_00017|M.leprae_Br4923 NGEPLNSVLKRTGRLSLRHALDMLEQTGRALQIAHAAGLVHRDVKPGNIL
MAB_0034c|M.abscessus_ATCC_199 HGEPLNSVLKRTGRLSLRHSLDMLEQTGRALQVAHAAGLVHRDVKPGNIL
Mflv_1389|M.gilvum_PYR-GCK PGRTLADVIE-QGPMPAPQVRRVLDEVLAALSVAHAAGVLHRDIKPANIL
Mvan_5401|M.vanbaalenii_PYR-1 PGRTLADLIA-QGPMAPAQVRAMLDDVLAALDVAHTAGVVHRDIKPGNIL
MSMEG_6206|M.smegmatis_MC2_155 SGATLADEIA-AGPLPQERVAELLDQILSALIAAHAAGIVHRDIKPGNIL
TH_2631|M.thermoresistible__bu DGVTLRDIVHTDGPMEPKRAIEVIADACQALNFSHQHGIIHRDVKPANIM
* .* . : * : : :: : ** :* *::***:**.**:
MMAR_0017|M.marinum_M ITPTG-QVKITDFGIAKAVD-AAPVTQTGMVMGTAQYIAPEQALGQDASP
MUL_0019|M.ulcerans_Agy99 ITPTG-QVKITDFGIAKAVD-AAPVTQTGMVMGTAQYIAPEQALGQDASP
MAV_0019|M.avium_104 ITPTG-QVKITDFGIAKAVD-AAPVTQTGMVMGTAQYIAPEQALGHDATP
Mb0015c|M.bovis_AF2122/97 ITPTG-QVKITDFGIAKAVD-AAPVTQTGMVMGTAQYIAPEQALGHDASP
Rv0015c|M.tuberculosis_H37Rv ITPTG-QVKITDFGIAKAVD-AAPVTQTGMVMGTAQYIAPEQALGHDASP
MLBr_00017|M.leprae_Br4923 ITPTG-QVKITDFGIAKAVD-AAPVTQTGMVMGTAQYIAPEQALGHDATP
MAB_0034c|M.abscessus_ATCC_199 ITPTG-QVKLTDFGIAKAVD-AAPVTQTGMVMGTAQYIAPEQALGHDATP
Mflv_1389|M.gilvum_PYR-GCK LTTAGDSVKVADFGIAKSAG--AAHTATGQIVGTLCYLSPERVMGAPASV
Mvan_5401|M.vanbaalenii_PYR-1 LSATGDRMQVADFGIAKSGG--AAHTMTGQIVGTLCYMSPERVSGAPASV
MSMEG_6206|M.smegmatis_MC2_155 IGHTG-QAKLADFGIAKTDG--VPHTQAGQILGTMAYLSPQRVTGQPAGF
TH_2631|M.thermoresistible__bu ISKTG-AVKVMDFGIARAIADSNSVTQTAAVIGTAQYLSPEQARGETVDA
: :* :: *****:: . * :. ::** *::*::. * .
MMAR_0017|M.marinum_M SSDVYSLGVVGYEAVSGKRPFTGDGALTVAMKHIKEPPPPLP---AELPP
MUL_0019|M.ulcerans_Agy99 SSDVYSLGVVGYEAVSGKRPFTGDGALTVAMKHIKEPPPPLP---AELPP
MAV_0019|M.avium_104 ASDVYSLGVVGYEVVSGKRPFSGDGALTVAMKHIKEPPPPLP---AELPP
Mb0015c|M.bovis_AF2122/97 ASDVYSLGVVGYEAVSGKRPFAGDGALTVAMKHIKEPPPPLP---PDLPP
Rv0015c|M.tuberculosis_H37Rv ASDVYSLGVVGYEAVSGKRPFAGDGALTVAMKHIKEPPPPLP---PDLPP
MLBr_00017|M.leprae_Br4923 ASDVYSLGVIGYEVVSGKRPFTGDGALTVAMKHIKEPPPPLP---ADLPP
MAB_0034c|M.abscessus_ATCC_199 ASDVYSLGVVGYEAVSGKRPFSGDGALTVAMKHIKETPSPLP---ADLPP
Mflv_1389|M.gilvum_PYR-GCK ADDLYAVGVMGYEALLGRRAFPQDNPAALARAIIDVPPPPLRALSVTADP
Mvan_5401|M.vanbaalenii_PYR-1 ADDLYAVGVMGYEALLARRAFPQESPVALVRAIVDAPPPALAGLTGGADP
MSMEG_6206|M.smegmatis_MC2_155 ADDLYAVGVVGYEALTGRRPFDRDNPAAMVRAILDDVPVPIASVRSDVAP
TH_2631|M.thermoresistible__bu RSDVYSLGCVLYEILTGEPPFVGDSPVAVAYQHVREDPVPPSRRNPAVSP
.*:*::* : ** : .. .* :.. ::. : * . *
MMAR_0017|M.marinum_M NVRELIEITLVKNPGLRYRSGGPFADAVAAVRAGRRPPRPSQSPPAGRAA
MUL_0019|M.ulcerans_Agy99 NVRELIEITLVKNPGLRYRSGGPFADAVAAVRAGRRPPRPSQSPPAGRAA
MAV_0019|M.avium_104 NVRELIEITLVKNPAMRYRSGGPFADAVAAVRAGRRPPRPSQSPPPGRAS
Mb0015c|M.bovis_AF2122/97 NVRELIEITLVKNPAMRYRSGGPFADAVAAVRAGRRPPRPSQTPPPGRAA
Rv0015c|M.tuberculosis_H37Rv NVRELIEITLVKNPAMRYRSGGPFADAVAAVRAGRRPPRPSQTPPPGRAA
MLBr_00017|M.leprae_Br4923 NVRELIEITLVKNPGMRYPSGGLFAEAVAAVRAGHRPPRPNQTPSSGRAS
MAB_0034c|M.abscessus_ATCC_199 NVRELIEITLVKNPQNRYTSGGPFADAVAAVRAGRRPPRPNQAPTISRAT
Mflv_1389|M.gilvum_PYR-GCK ALVAVIDRAMSRDPRLRFGSAPQMRAALAGDPLALNNAPPPAPPPRAGTR
Mvan_5401|M.vanbaalenii_PYR-1 VLAAVIDRAMSRDPRRRFGSAAQMRAALAGDVTALSTAAEPRP----LTK
MSMEG_6206|M.smegmatis_MC2_155 GLIHVIDMAMAREVAFRYPDAATMRAALTGAEPPTQRVG-RRQ-----TK
TH_2631|M.thermoresistible__bu ELDAVVLKALAKNPDNRYQTAAEMRADLVRVHSGEQPEAPKVLTDAERTS
: :: :: :: *: . : :. :
MMAR_0017|M.marinum_M PAAIPSSAPARIAPAPTTRTTTPR-RTRPATGGHRPPPA-RRTFSSGQRA
MUL_0019|M.ulcerans_Agy99 PAAIPSSAPARIAPAPTTRTTTPR-RTRPATGGHRPPPS-RRTFSSGQRA
MAV_0019|M.avium_104 PAAIPSSPTTRAAAVSSGRTAAPR-RTRPATGGHRPPPA-RRTFSSGQRA
Mb0015c|M.bovis_AF2122/97 PAAIPSGTTARVAANSAGRTAASR-RSRPATGGHRPP---RRTFSSGQRA
Rv0015c|M.tuberculosis_H37Rv PAAIPSGTTARVAANSAGRTAASR-RSRPATGGHRPP---RRTFSSGQRA
MLBr_00017|M.leprae_Br4923 PTTIPSSTQARAAVACGTKTPAPR-RSRPSTSGNRPPPA-RNTFSSGQRA
MAB_0034c|M.abscessus_ATCC_199 PAAIPPATQVRPAVGAPSRVAPPTSRTRQPTGSHRPPPPRRGTFSPGQKA
Mflv_1389|M.gilvum_PYR-GCK VLAQPLPPSAGFTSAPVGYPTAAPP-PPTRAR---MYLMAVAAVL-AFTV
Mvan_5401|M.vanbaalenii_PYR-1 VLAAPLPPSA-------AYPAPAPRRPATRAR---RYVVGAAAVMTAFVI
MSMEG_6206|M.smegmatis_MC2_155 VLAAPLPPIP---------PPPVLPSPRHWRT---RLILAVLGIV-STLI
TH_2631|M.thermoresistible__bu LLASGPAHDRTQPTDAVIDPVIPRYTGSDRGGSVGRWVAAVAVLAVLTVV
: .
MMAR_0017|M.marinum_M LLWAAGVLGALAIIIAVLIVIN-----------------SRAD-SQQQPP
MUL_0019|M.ulcerans_Agy99 LLWAAGVLGALAIIIAVLIVIN-----------------SRAD-SQQQSP
MAV_0019|M.avium_104 LLWAAGVLGALAIIIAVLIVIN-----------------SRAD-QQQQ--
Mb0015c|M.bovis_AF2122/97 LLWAAGVLGALAIIIAVLLVIK-----------------APGDNSPQQAP
Rv0015c|M.tuberculosis_H37Rv LLWAAGVLGALAIIIAVLLVIK-----------------APGDNSPQQAP
MLBr_00017|M.leprae_Br4923 LLWAAGMLGALAIIIAVLIVIN-----------------SYAGNEQHQPP
MAB_0034c|M.abscessus_ATCC_199 LMWAAAVLGIFAIIIAALILLQ-----------------ASEKKNQTPSI
Mflv_1389|M.gilvum_PYR-GCK AALALALDPFSSPPP-MQTVG----------------TSTTAPPPPTSVP
Mvan_5401|M.vanbaalenii_PYR-1 SALALAMDPFSSSAPRPEPVS----------------TSTSAPSPTTPPP
MSMEG_6206|M.smegmatis_MC2_155 LVLVLALARGSSGLTSPTSTP----------------TATTVPPATSAVV
TH_2631|M.thermoresistible__bu VTVAINMIGGNPRNVQVPDVSGQASADAIAALQNRGFKTRTQQQPDNSVP
. : .
MMAR_0017|M.marinum_M PPTVTQTTTAHQTPTGQGPRLN----------------------------
MUL_0019|M.ulcerans_Agy99 PPTVTQTTTAHQTPTGQGPRLN----------------------------
MAV_0019|M.avium_104 PPTVTDTG----TPPASAPPPT----------------------------
Mb0015c|M.bovis_AF2122/97 TPTVTTTG----NPPASN--------------------------------
Rv0015c|M.tuberculosis_H37Rv TPTVTTTG----NPPASN--------------------------------
MLBr_00017|M.leprae_Br4923 TPTVTDTG----TPPATKT-------------------------------
MAB_0034c|M.abscessus_ATCC_199 PATVSGSE----TPSSPGS-------------------------------
Mflv_1389|M.gilvum_PYR-GCK PPTSAAPLPPPIVPVTQ---------------------------------
Mvan_5401|M.vanbaalenii_PYR-1 PPITTPPPSP--APAVT---------------------------------
MSMEG_6206|M.smegmatis_MC2_155 PPPVPSTPVPAAVPEEP---------------------------------
TH_2631|M.thermoresistible__bu PDRVISTDPPADTSVSAGDEITVNVSTGPQQREIPDVSSLTYAEAVRRLS
. . .
MMAR_0017|M.marinum_M ----------------WTEGKAIGNPGLQSNQPDLVDDAGRNRPIGNR-G
MUL_0019|M.ulcerans_Agy99 ----------------WTDGQSIGNPGLQSNQPDLVDDAGRNRPIGNR-D
MAV_0019|M.avium_104 ----------------KTPSGSGAHPGLRLDWP----DDGTIGASGFRDG
Mb0015c|M.bovis_AF2122/97 ------------------TGGTDASP--RLNWT----ERGETRHSGLQ--
Rv0015c|M.tuberculosis_H37Rv ------------------TGGTDASP--RLNWT----ERGETRHSGLQ--
MLBr_00017|M.leprae_Br4923 -----------------LSGFPAAYCEYRVNWT----NHKEISNSGLP--
MAB_0034c|M.abscessus_ATCC_199 -----------------ASPGALAPANIVALLP----DAEPDPIPGTK--
Mflv_1389|M.gilvum_PYR-GCK ------------------APP---PDNGPKKGPDKKGEKGN---NGRGNG
Mvan_5401|M.vanbaalenii_PYR-1 ------------------AAT---QQPEPQQGRDDKGPRGNGRGNGKGKG
MSMEG_6206|M.smegmatis_MC2_155 ------------------RGPGRGPGPGPGPGKGKGHEKGHGPKGPKGPP
TH_2631|M.thermoresistible__bu AAGFEKFRQSESPSTPELRGKVVATNPPANQTAAITNEITVVVGTGPATR
MMAR_0017|M.marinum_M SATSWNVTVTPQHLAALARFEMQR--------------------------
MUL_0019|M.ulcerans_Agy99 SATSWNVTVTPQHRAALARFEMQR--------------------------
MAV_0019|M.avium_104 PARHWTSQ------------------------------------------
Mb0015c|M.bovis_AF2122/97 ---SWVVPPTPHSRASLARYEIAQ--------------------------
Rv0015c|M.tuberculosis_H37Rv ---SWVVPPTPHSRASLARYEIAQ--------------------------
MLBr_00017|M.leprae_Br4923 ---KQAARAQLAGAIDISPVAGQT--------------------------
MAB_0034c|M.abscessus_ATCC_199 ---------SP---------------------------------------
Mflv_1389|M.gilvum_PYR-GCK NKPD----------------------------------------------
Mvan_5401|M.vanbaalenii_PYR-1 EKKD----------------------------------------------
MSMEG_6206|M.smegmatis_MC2_155 HP------------------------------------------------
TH_2631|M.thermoresistible__bu EVPDITGQTVELAERNLSVYGFTKFTQSAVDSTAPAGQVVGTNPPAGTTV
MMAR_0017|M.marinum_M --------------------------------------------------
MUL_0019|M.ulcerans_Agy99 --------------------------------------------------
MAV_0019|M.avium_104 --------------------------------------------------
Mb0015c|M.bovis_AF2122/97 --------------------------------------------------
Rv0015c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00017|M.leprae_Br4923 --------------------------------------------------
MAB_0034c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1389|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5401|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6206|M.smegmatis_MC2_155 --------------------------------------------------
TH_2631|M.thermoresistible__bu SLDSVIELEVSKGNQFVMPDLIGMFWTDAEPRLRALGWTGVLDKGADIPN
MMAR_0017|M.marinum_M -------------------------------
MUL_0019|M.ulcerans_Agy99 -------------------------------
MAV_0019|M.avium_104 -------------------------------
Mb0015c|M.bovis_AF2122/97 -------------------------------
Rv0015c|M.tuberculosis_H37Rv -------------------------------
MLBr_00017|M.leprae_Br4923 -------------------------------
MAB_0034c|M.abscessus_ATCC_199 -------------------------------
Mflv_1389|M.gilvum_PYR-GCK -------------------------------
Mvan_5401|M.vanbaalenii_PYR-1 -------------------------------
MSMEG_6206|M.smegmatis_MC2_155 -------------------------------
TH_2631|M.thermoresistible__bu SGQRANAVVRQNPSPGAGVNYDARITLNFAS