For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LEGMQAPEVLGGRYRLGPVLGRGGMAEVRDAWDQRLGRPVAVKLLYPSVSTQPDTRRRFATEARAAAALN HPHVVAVHDTGVHEGRHYIVLERLPGQNLADLLARHGPLPVEQVRAILRDVLSALSAAHLSGVLHRDIKP ANILFTEFGGVKIADFGVAKSPDTPQTLTNRVFGTMAYLPSDRIAGRPATPSDDLYALGVVAYEALTGRR AYPQDNLSALADAIAAGRLAPLTSVRPDVDPALAATIERAIARDPRWRFATAEQMRRSLDPPPPRAGRAG AVLAVAAILLIVALVALLVVA
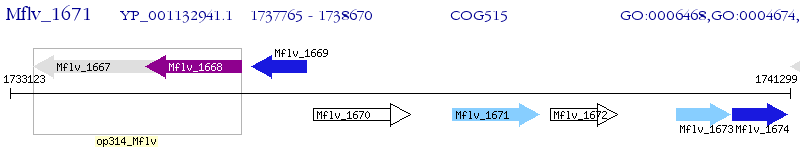
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1671 | - | - | 100% (301) | protein kinase |
| M. gilvum PYR-GCK | Mflv_1389 | - | 2e-83 | 56.89% (283) | protein kinase |
| M. gilvum PYR-GCK | Mflv_0812 | - | 8e-48 | 38.83% (273) | protein kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0015c | pknA | 1e-47 | 39.40% (302) | transmembrane serine/threonine-protein kinase A PKNA |
| M. tuberculosis H37Rv | Rv0015c | pknA | 1e-47 | 39.40% (302) | transmembrane serine/threonine-protein kinase A PKNA |
| M. leprae Br4923 | MLBr_00016 | pknB | 7e-47 | 39.34% (272) | putative serine/threonine protein kinase |
| M. abscessus ATCC 19977 | MAB_0033c | - | 4e-47 | 37.96% (274) | serine/threonine-protein kinase PknB |
| M. marinum M | MMAR_0017 | pknA | 6e-47 | 41.11% (287) | serine/threonine-protein kinase a PknA |
| M. avium 104 | MAV_2318 | - | 6e-47 | 39.03% (269) | serine-threonine protein kinase |
| M. smegmatis MC2 155 | MSMEG_6206 | - | 5e-79 | 55.68% (273) | serine/threonine protein kinase PksC |
| M. thermoresistible (build 8) | TH_2631 | - | 8e-48 | 39.71% (272) | PUTATIVE serine/threonine protein kinase |
| M. ulcerans Agy99 | MUL_0019 | pknA | 4e-47 | 41.11% (287) | serine/threonine-protein kinase a PknA |
| M. vanbaalenii PYR-1 | Mvan_5082 | - | 1e-140 | 82.83% (297) | protein kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1671|M.gilvum_PYR-GCK ------------------------MEGMQAPEVLGGRYRLGPVLGRGGMA
Mvan_5082|M.vanbaalenii_PYR-1 ---------------------------MQAPELIGGRYELRGVLGRGGMA
MSMEG_6206|M.smegmatis_MC2_155 -----------------------MGSAERFQRLLGDRYELRGVLGRGGMA
Mb0015c|M.bovis_AF2122/97 -------------------------MSPRVGVTLSGRYRLQRLIATGGMG
Rv0015c|M.tuberculosis_H37Rv -------------------------MSPRVGVTLSGRYRLQRLIATGGMG
MMAR_0017|M.marinum_M -------------------------MSPRVGMTLSGRYRLQRLIATGGMG
MUL_0019|M.ulcerans_Agy99 -------------------------MSPIVGMTLSGRYRLQRLIATGGMG
MLBr_00016|M.leprae_Br4923 ---------------------------MTTPPHLSDRYELGDILGFGGMS
TH_2631|M.thermoresistible__bu ---------------------------MTTPQHLSDRYELGEILGFGGMS
MAB_0033c|M.abscessus_ATCC_199 ---------------------------MTTPQHLSDRYELGEILGYGGMS
MAV_2318|M.avium_104 MAQAESPGSSSGRPGPHRRQAQAESPGSFAGTLLDGRYLIESKIASGGTS
:..** : :. ** .
Mflv_1671|M.gilvum_PYR-GCK EVRDAWDQRLGRPVAVKLLYPSVSTQPDTRRRFATEARAAAALNHPHVVA
Mvan_5082|M.vanbaalenii_PYR-1 EVRDAWDKRLGRPVAVKLLYPSVSAHPDTRRRFAIEARAAAALNHPHVVA
MSMEG_6206|M.smegmatis_MC2_155 EVRDGWDTRLARPVAIKLLYPALAADADLRARFEREARAAAALTHPHVVA
Mb0015c|M.bovis_AF2122/97 QVWEAVDNRLGRRVAVKVLKSEFSSDPEFIERFRAEARTTAMLNHPGIAS
Rv0015c|M.tuberculosis_H37Rv QVWEAVDNRLGRRVAVKVLKSEFSSDPEFIERFRAEARTTAMLNHPGIAS
MMAR_0017|M.marinum_M QVWEAVDSRLGRRVAVKVLKQEFSSDPEFIERFRAEARTTAMLNHPGIAA
MUL_0019|M.ulcerans_Agy99 QVWEAVDSRLGRRVAVKVLKQEFSSDPEFIERFRAEARTTAMLNHPGIAA
MLBr_00016|M.leprae_Br4923 EVHLARDIRLHRDVAVKVLRADLARDPSFYLRFRREAQNAAALNHPSIVA
TH_2631|M.thermoresistible__bu EVHLARDLRLHRDVAVKVLRADLARDPSFYLRFRREAQNAAALNHPAIVA
MAB_0033c|M.abscessus_ATCC_199 EVHLARDLRLNRDVAIKVLRADLARDPSFYLRFRREAQNAAALNHPAIVA
MAV_2318|M.avium_104 TVYRGVDTRLDRPVAVKVMDPRYAGDEQFLTRFQREARAVARLKDPGLVA
* . * ** * **:*:: : . . ** **: .* *..* :.:
Mflv_1671|M.gilvum_PYR-GCK VHDTGVHEGR-----HYIVLERLPGQNLADLLARHGPLPVEQVRAILRDV
Mvan_5082|M.vanbaalenii_PYR-1 VHDSGVHDGR-----HYIVLERLPGQSLADVLAHHGPLPVDHVRAIMRDV
MSMEG_6206|M.smegmatis_MC2_155 VHDCGEHAGT-----PYIVMERLSGATLADEIA-AGPLPQERVAELLDQI
Mb0015c|M.bovis_AF2122/97 VHDYGESQMNGEGRTAYLVMELVNGEPLNSVLKRTGRLSLRHALDMLEQT
Rv0015c|M.tuberculosis_H37Rv VHDYGESQMNGEGRTAYLVMELVNGEPLNSVLKRTGRLSLRHALDMLEQT
MMAR_0017|M.marinum_M VHDYGESQMNGEGRTAYLVMELVNGEPLNSVLKRTGRLSLRHALDMLEQT
MUL_0019|M.ulcerans_Agy99 VHDYGESQMNGEGRTAYLVMELVNGEPLNSVLKRTGRLSLRHALDMLEQT
MLBr_00016|M.leprae_Br4923 VYDTGEAETS-AGPLPYIVMEYVDGATLRDIVHTDGPMPPQQAIEIVADA
TH_2631|M.thermoresistible__bu VYDTGEAETP-TGPLPYIVMEYVDGVTLRDIVHTDGPMEPKRAIEVIADA
MAB_0033c|M.abscessus_ATCC_199 VYDTGEAETA-TGPLPYIVMEYVDGITLRDIVKDDGPMPIRRAIEVIADS
MAV_2318|M.avium_104 VYDQGLDARH-----PFLVMELIEGGTLRELLGERGPMPPYAVAAVLRPV
*:* * ::*:* : * * . : * : . ::
Mflv_1671|M.gilvum_PYR-GCK LSALSAAHLSGVLHRDIKPANILFTEFGGVKIADFGVAKSPDTP---QTL
Mvan_5082|M.vanbaalenii_PYR-1 LSALRAAHTSGVLHRDIKPANILFTHFGGVKIADFGVAKSADTP---QTL
MSMEG_6206|M.smegmatis_MC2_155 LSALIAAHAAGIVHRDIKPGNILIGHTGQAKLADFGIAKTDGVP---HTQ
Mb0015c|M.bovis_AF2122/97 GRALQIAHAAGLVHRDVKPGNILITPTGQVKITDFGIAKAVDAA--PVTQ
Rv0015c|M.tuberculosis_H37Rv GRALQIAHAAGLVHRDVKPGNILITPTGQVKITDFGIAKAVDAA--PVTQ
MMAR_0017|M.marinum_M GRALQVAHAAGLVHRDVKPGNILITPTGQVKITDFGIAKAVDAA--PVTQ
MUL_0019|M.ulcerans_Agy99 GRALQVAHAAGLVHRDVKPGNILITPTGQVKITDFGIAKAVDAA--PVTQ
MLBr_00016|M.leprae_Br4923 CQALNFSHQNGIIHRDVKPANIMISATNAVKVMDFGIARAIADS-TSVTQ
TH_2631|M.thermoresistible__bu CQALNFSHQHGIIHRDVKPANIMISKTGAVKVMDFGIARAIADS-NSVTQ
MAB_0033c|M.abscessus_ATCC_199 CQALNFSHQHGIIHRDVKPANIMISKTGAVKVMDFGIARAVSDAGNSVTQ
MAV_2318|M.avium_104 LGGLAAAHRAGLVHRDVKPENVLISDDGEVKIADFGLVRAVAAAG--ITS
.* :* *::***:** *::: . .*: ***:.:: . *
Mflv_1671|M.gilvum_PYR-GCK TNRVFGTMAYLPSDRIAGRPATPSDDLYALGVVAYEALTGRRAYPQDNLS
Mvan_5082|M.vanbaalenii_PYR-1 TNRVFGTMAYLPADRIAGRPATPSDDLYALGVVAYEALTARRAYPQENLT
MSMEG_6206|M.smegmatis_MC2_155 AGQILGTMAYLSPQRVTGQPAGFADDLYAVGVVGYEALTGRRPFDRDNPA
Mb0015c|M.bovis_AF2122/97 TGMVMGTAQYIAPEQALGHDASPASDVYSLGVVGYEAVSGKRPFAGDGAL
Rv0015c|M.tuberculosis_H37Rv TGMVMGTAQYIAPEQALGHDASPASDVYSLGVVGYEAVSGKRPFAGDGAL
MMAR_0017|M.marinum_M TGMVMGTAQYIAPEQALGQDASPSSDVYSLGVVGYEAVSGKRPFTGDGAL
MUL_0019|M.ulcerans_Agy99 TGMVMGTAQYIAPEQALGQDASPSSDVYSLGVVGYEAVSGKRPFTGDGAL
MLBr_00016|M.leprae_Br4923 TAAVIGTAQYLSPEQARGDSVDARSDVYSLGCVLYEILTGEPPFIGDSPV
TH_2631|M.thermoresistible__bu TAAVIGTAQYLSPEQARGETVDARSDVYSLGCVLYEILTGEPPFVGDSPV
MAB_0033c|M.abscessus_ATCC_199 TAAVIGTAQYLSPEQARGETVDARSDVYSLGCVLYELVTGEPPFVGDSPV
MAV_2318|M.avium_104 ASVILGTAAYLSPEQVRDGAATPRSDVYAAGILAYELLTGRTPFTGDSTL
: ::** *:..:: . . .*:*: * : ** ::.. .: :.
Mflv_1671|M.gilvum_PYR-GCK ALADAIAAGRLAPLTSVRPDVDPALAATIERAIARDPRWRFATAEQMRRS
Mvan_5082|M.vanbaalenii_PYR-1 ALADAIAAGHLAPVATLRPDVDPALATTIERSMARDPRWRFATADQMLAC
MSMEG_6206|M.smegmatis_MC2_155 AMVRAILDDVPVPIASVRSDVAPGLIHVIDMAMAREVAFRYPDAATMRAA
Mb0015c|M.bovis_AF2122/97 TVAMKHIKE---PPPPLPPDLPPNVRELIEITLVKNPAMRYRSGGPFADA
Rv0015c|M.tuberculosis_H37Rv TVAMKHIKE---PPPPLPPDLPPNVRELIEITLVKNPAMRYRSGGPFADA
MMAR_0017|M.marinum_M TVAMKHIKE---PPPPLPAELPPNVRELIEITLVKNPGLRYRSGGPFADA
MUL_0019|M.ulcerans_Agy99 TVAMKHIKE---PPPPLPAELPPNVRELIEITLVKNPGLRYRSGGPFADA
MLBr_00016|M.leprae_Br4923 SVAYQHVREDPIPPSQRHEGISVDLDAVVLKALAKNPENRYQTAAEMRAD
TH_2631|M.thermoresistible__bu AVAYQHVREDPVPPSRRNPAVSPELDAVVLKALAKNPDNRYQTAAEMRAD
MAB_0033c|M.abscessus_ATCC_199 AVAYQHVREDPVAPSKHNPEIPPGLDSVVLKALSKNPDNRYQNAAEMRAD
MAV_2318|M.avium_104 AIAYRRLDADVPPPSAAIDGVPAQFDDFVQRATARDPADRYADAVEMGAD
::. . . : . : : :: *: . :
Mflv_1671|M.gilvum_PYR-GCK LD------------------------------------------------
Mvan_5082|M.vanbaalenii_PYR-1 LD------------------------------------------------
MSMEG_6206|M.smegmatis_MC2_155 LT------------------------------------------------
Mb0015c|M.bovis_AF2122/97 VAAVRAGRRP----------------------------------------
Rv0015c|M.tuberculosis_H37Rv VAAVRAGRRP----------------------------------------
MMAR_0017|M.marinum_M VAAVRAGRRP----------------------------------------
MUL_0019|M.ulcerans_Agy99 VAAVRAGRRP----------------------------------------
MLBr_00016|M.leprae_Br4923 LIRVHSGQPPEAPKVLTDADRSCLLSSGAGNFGVPRTDALSR---QSLDE
TH_2631|M.thermoresistible__bu LVRVHSGEQPEAPKVLTDAERTSLLASGPAHDRTQPTDAVIDPVIPRYTG
MAB_0033c|M.abscessus_ATCC_199 LMRVYQGEQPEAPKVLTDAERSTMLSTPANTRAFPASGPQTGPLPSQYEE
MAV_2318|M.avium_104 LDAIADELALPG--------------------------------------
:
Mflv_1671|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5082|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6206|M.smegmatis_MC2_155 --------------------------------------------------
Mb0015c|M.bovis_AF2122/97 --------------------------------------------------
Rv0015c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0017|M.marinum_M --------------------------------------------------
MUL_0019|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00016|M.leprae_Br4923 TESDGSIGRWVAVVAVLAVLTIAIVAAFNTFGGNTRDVQVPDVRGQVSAD
TH_2631|M.thermoresistible__bu SDRGGSVGRWVAAVAVLAVLTVVVTVAINMIGGNPRNVQVPDVSGQASAD
MAB_0033c|M.abscessus_ATCC_199 PRRN-PLSRWLVGLAALVVLTIIVTFAIILGNGPSRDVQVPDVTGKSQSD
MAV_2318|M.avium_104 --------------------------------------------------
Mflv_1671|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5082|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6206|M.smegmatis_MC2_155 --------------------------------------------------
Mb0015c|M.bovis_AF2122/97 -------------------------------PRPSQTPPPGRAAPAAIPS
Rv0015c|M.tuberculosis_H37Rv -------------------------------PRPSQTPPPGRAAPAAIPS
MMAR_0017|M.marinum_M -------------------------------PRPSQSPPAGRAAPAAIPS
MUL_0019|M.ulcerans_Agy99 -------------------------------PRPSQSPPAGRAAPAAIPS
MLBr_00016|M.leprae_Br4923 AISALQNRGFKTRTLQKPDSTIPPDHVISTEPGANASVGAGDEITINVST
TH_2631|M.thermoresistible__bu AIAALQNRGFKTRTQQQPDNSVPPDRVISTDPPADTSVSAGDEITVNVST
MAB_0033c|M.abscessus_ATCC_199 AVAALQNLGFKTKVQKNTSSKVAPEKVIDTDPAKGSTVSAGAEVTVNVSI
MAV_2318|M.avium_104 --------------------------------------------------
Mflv_1671|M.gilvum_PYR-GCK ------PPPPRAGRAG-----------------------------AVLAV
Mvan_5082|M.vanbaalenii_PYR-1 ------GPAQRPQRTG-----------------------------VVLAA
MSMEG_6206|M.smegmatis_MC2_155 ------GAEPPTQRVGRRQTKVLAAPLPPIPPPPVLPSPRHWRTRLILAV
Mb0015c|M.bovis_AF2122/97 GTTARVAANSAGRTAASRRSRPATGGHRPP--RRTFSSGQRALLWAAGVL
Rv0015c|M.tuberculosis_H37Rv GTTARVAANSAGRTAASRRSRPATGGHRPP--RRTFSSGQRALLWAAGVL
MMAR_0017|M.marinum_M SAPARIAPAPTTRTTTPRRTRPATGGHRPPPARRTFSSGQRALLWAAGVL
MUL_0019|M.ulcerans_Agy99 SAPARIAPAPTTRTTTPRRTRPATGGHRPPPSRRTFSSGQRALLWAAGVL
MLBr_00016|M.leprae_Br4923 GPEQREVPDVSSLNYTDAVKKLTSSGFK-SFKQANSPSTPELLGKVIGTN
TH_2631|M.thermoresistible__bu GPQQREIPDVSSLTYAEAVRRLSAAGFE-KFRQSESPSTPELRGKVVATN
MAB_0033c|M.abscessus_ATCC_199 GPENQQVPDCHGLSFGDAVKKLQAAGFKGTFTQSETKSLPEDKGRVMATN
MAV_2318|M.avium_104 ----FRVPAPRNSALHRSAALHREAGRRAPGAEPPARHPTRHLTRGPEEW
.
Mflv_1671|M.gilvum_PYR-GCK AAILLIVALVALLVVA----------------------------------
Mvan_5082|M.vanbaalenii_PYR-1 AALLAVFVLAALLVVV----------------------------------
MSMEG_6206|M.smegmatis_MC2_155 LGIVSTLILVLVLALARGSSGLTSPTSTPTAT------------------
Mb0015c|M.bovis_AF2122/97 GALAIIIAVLLVIKAPGDNSPQQAPTPTVTTT------------------
Rv0015c|M.tuberculosis_H37Rv GALAIIIAVLLVIKAPGDNSPQQAPTPTVTTT------------------
MMAR_0017|M.marinum_M GALAIIIAVLIVINSRAD-SQQQPPPPTVTQT------------------
MUL_0019|M.ulcerans_Agy99 GALAIIIAVLIVINSRAD-SQQQSPPPTVTQT------------------
MLBr_00016|M.leprae_Br4923 PSANQTSAITNVITIIVGSGPETKQIPDVTGQIVEIAQKNLNVYGFTK-F
TH_2631|M.thermoresistible__bu PPANQTAAITNEITVVVGTGPATREVPDITGQTVELAERNLSVYGFTK-F
MAB_0033c|M.abscessus_ATCC_199 PPAGQYSAVTNNITIVVGKGPGEKPVPDLGGQNIDVAIKNLPTYGFTAPP
MAV_2318|M.avium_104 PQPEPPAHVGAEPDDDEDDYEYQSVTGEFAGIPIS---------------
:
Mflv_1671|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5082|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6206|M.smegmatis_MC2_155 ----------------TVPPATSAVVPPPVPSTPVPAAVPEEPRGPGRGP
Mb0015c|M.bovis_AF2122/97 ----------------GNPPASNTGGTDASPRLNWTERGETRHSGLQS--
Rv0015c|M.tuberculosis_H37Rv ----------------GNPPASNTGGTDASPRLNWTERGETRHSGLQS--
MMAR_0017|M.marinum_M ----------------TTAHQTPTG---QGPRLNWTEGKAIGNPGLQSNQ
MUL_0019|M.ulcerans_Agy99 ----------------TTAHQTPTG---QGPRLNWTDGQSIGNPGLQSNQ
MLBr_00016|M.leprae_Br4923 SQASVDSPRPTGEVIGTNPPKDATVPVDSVIELQVSKGNQFVMPDLSGMF
TH_2631|M.thermoresistible__bu TQSAVDSTAPAGQVVGTNPPAGTTVSLDSVIELEVSKGNQFVMPDLIGMF
MAB_0033c|M.abscessus_ATCC_199 TVIPVPSPESKGQILSTNPPANTMFPEDGVIEIRVSNGGQFRMPNIMGKF
MAV_2318|M.avium_104 ------------EFVWARQHNRRMVLVWLALVLAVTGMVATAAWTIGRNL
Mflv_1671|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5082|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6206|M.smegmatis_MC2_155 GPGPGPGKGKGH--------------------EKGHGPKGPKGPPHP---
Mb0015c|M.bovis_AF2122/97 -------------------------------WVVPPTPHSRASLARYEIA
Rv0015c|M.tuberculosis_H37Rv -------------------------------WVVPPTPHSRASLARYEIA
MMAR_0017|M.marinum_M PDLVDDAGRNR----------PIGNRGSATSWNVTVTPQHLAALARFEMQ
MUL_0019|M.ulcerans_Agy99 PDLVDDAGRNR----------PIGNRDSATSWNVTVTPQHRAALARFEMQ
MLBr_00016|M.leprae_Br4923 WADAEPRLRALGWTGVLDKGPDVDAGGSQHNRVAYQNPPAGAGVNRDGII
TH_2631|M.thermoresistible__bu WTDAEPRLRALGWTGVLDKGADIPNSGQRANAVVRQNPSPGAGVNYDARI
MAB_0033c|M.abscessus_ATCC_199 WTDAEPMLYQAGWQGGGLSTEDVPSDGNNRARVVFQDPPAGAGVGYNGPI
MAV_2318|M.avium_104 NGLF----------------------------------------------
Mflv_1671|M.gilvum_PYR-GCK -------
Mvan_5082|M.vanbaalenii_PYR-1 -------
MSMEG_6206|M.smegmatis_MC2_155 -------
Mb0015c|M.bovis_AF2122/97 Q------
Rv0015c|M.tuberculosis_H37Rv Q------
MMAR_0017|M.marinum_M R------
MUL_0019|M.ulcerans_Agy99 R------
MLBr_00016|M.leprae_Br4923 TLKFGQ-
TH_2631|M.thermoresistible__bu TLNFAS-
MAB_0033c|M.abscessus_ATCC_199 KLRFGAG
MAV_2318|M.avium_104 -------