For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SPIVGMTLSGRYRLQRLIATGGMGQVWEAVDSRLGRRVAVKVLKQEFSSDPEFIERFRAEARTTAMLNHP GIAAVHDYGESQMNGEGRTAYLVMELVNGEPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRD VKPGNILITPTGQVKITDFGIAKAVDAAPVTQTGMVMGTAQYIAPEQALGQDASPSSDVYSLGVVGYEAV SGKRPFTGDGALTVAMKHIKEPPPPLPAELPPNVRELIEITLVKNPGLRYRSGGPFADAVAAVRAGRRPP RPSQSPPAGRAAPAAIPSSAPARIAPAPTTRTTTPRRTRPATGGHRPPPSRRTFSSGQRALLWAAGVLGA LAIIIAVLIVINSRADSQQQSPPPTVTQTTTAHQTPTGQGPRLNWTDGQSIGNPGLQSNQPDLVDDAGRN RPIGNRDSATSWNVTVTPQHRAALARFEMQR*
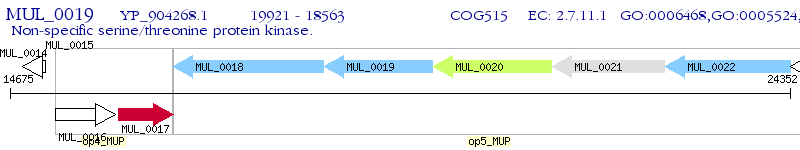
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0019 | pknA | - | 100% (452) | serine/threonine-protein kinase a PknA |
| M. ulcerans Agy99 | MUL_0018 | pknB | 6e-61 | 38.58% (381) | serine/threonine-protein kinase B PknB |
| M. ulcerans Agy99 | MUL_3519 | pknL | 6e-53 | 42.08% (259) | transmembrane serine/threonine-protein kinase L PknL |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0015c | pknA | 0.0 | 79.96% (454) | transmembrane serine/threonine-protein kinase A PKNA |
| M. gilvum PYR-GCK | Mflv_0811 | - | 1e-176 | 74.04% (443) | protein kinase |
| M. tuberculosis H37Rv | Rv0015c | pknA | 0.0 | 79.96% (454) | transmembrane serine/threonine-protein kinase A PKNA |
| M. leprae Br4923 | MLBr_00017 | pknA | 0.0 | 82.17% (415) | putative serine/threonine protein kinase |
| M. abscessus ATCC 19977 | MAB_0034c | - | 1e-170 | 77.35% (393) | putative serine/threonine-protein kinase PknA |
| M. marinum M | MMAR_0017 | pknA | 0.0 | 98.23% (452) | serine/threonine-protein kinase a PknA |
| M. avium 104 | MAV_0019 | - | 0.0 | 86.20% (413) | serine/threonine protein kinases Drp72 |
| M. smegmatis MC2 155 | MSMEG_0030 | pknA | 0.0 | 82.53% (395) | serine/threonine protein kinase PknA |
| M. thermoresistible (build 8) | TH_2631 | - | 9e-63 | 35.91% (440) | PUTATIVE serine/threonine protein kinase |
| M. vanbaalenii PYR-1 | Mvan_0024 | - | 1e-177 | 80.21% (389) | protein kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0811|M.gilvum_PYR-GCK MSPRVGVTLSGRYRLQRLIATGGMGQVWEGVDSRLGRRVAIKVLKAEYST
Mvan_0024|M.vanbaalenii_PYR-1 MSPRVGVTLSGRYRLQRLIATGGMGQVWEGVDSRLGRRVAIKVLKAEYST
MSMEG_0030|M.smegmatis_MC2_155 MSPRVGVTLSGRYRLQRLIATGGMGQVWEAVDSRLGRRVAVKVLKAEFST
MAB_0034c|M.abscessus_ATCC_199 MTVRVGETLSGRYRLQRLIATGGMGQVWEGTDSRLGRKVAVKVLKAEFSS
Mb0015c|M.bovis_AF2122/97 MSPRVGVTLSGRYRLQRLIATGGMGQVWEAVDNRLGRRVAVKVLKSEFSS
Rv0015c|M.tuberculosis_H37Rv MSPRVGVTLSGRYRLQRLIATGGMGQVWEAVDNRLGRRVAVKVLKSEFSS
MUL_0019|M.ulcerans_Agy99 MSPIVGMTLSGRYRLQRLIATGGMGQVWEAVDSRLGRRVAVKVLKQEFSS
MMAR_0017|M.marinum_M MSPRVGMTLSGRYRLQRLIATGGMGQVWEAVDSRLGRRVAVKVLKQEFSS
MAV_0019|M.avium_104 ------MTLSGRYRLQRLIATGGMGQVWEAVDNRLGRRVAVKVLKQEFSQ
MLBr_00017|M.leprae_Br4923 MSPRIGVMLSGRYRLHRLIATGGMGQVWEAVDSRLGRRVAVKVLKGEFSS
TH_2631|M.thermoresistible__bu --MTTPQHLSDRYELGEILGFGGMSEVHLARDLRLHRDVAVKVLRADLAR
**.**.* .::. ***.:* . * ** * **:***: : :
Mflv_0811|M.gilvum_PYR-GCK DSEFVERFRAEARTVAMLNHPGIAGVYDYGETDIDGEGRTAYLVMELVNG
Mvan_0024|M.vanbaalenii_PYR-1 DAEFVERFRAEARTVAMLNHPGIAGVYDYGETDIDGEGRTAYLVMELVNG
MSMEG_0030|M.smegmatis_MC2_155 DAEFVERFRAEARTVAMLNHPGIASVYDYGETELDGEGRTAYLVMELING
MAB_0034c|M.abscessus_ATCC_199 DPEFIERFRAEARTVAMLNHPGIASVYDYGETDIDSEGRTAYLVMELIHG
Mb0015c|M.bovis_AF2122/97 DPEFIERFRAEARTTAMLNHPGIASVHDYGESQMNGEGRTAYLVMELVNG
Rv0015c|M.tuberculosis_H37Rv DPEFIERFRAEARTTAMLNHPGIASVHDYGESQMNGEGRTAYLVMELVNG
MUL_0019|M.ulcerans_Agy99 DPEFIERFRAEARTTAMLNHPGIAAVHDYGESQMNGEGRTAYLVMELVNG
MMAR_0017|M.marinum_M DPEFIERFRAEARTTAMLNHPGIAAVHDYGESQMNGEGRTAYLVMELVNG
MAV_0019|M.avium_104 DPEFIERFRAEARTTAMLNHPGIAAVHDYGESQLDGEGRTAYLVMELVNG
MLBr_00017|M.leprae_Br4923 DPEFIERFRAEARTTAMLNHPGIASVHDYGESHMDGEGRTAYLVMELVNG
TH_2631|M.thermoresistible__bu DPSFYLRFRREAQNAAALNHPAIVAVYDTGEAETP-TGPLPYIVMEYVDG
*..* *** **:..* ****.*..*:* **:. * .*:*** :.*
Mflv_0811|M.gilvum_PYR-GCK EPLNSVIKRTGRLSLRHALDMLEQTGRALQVAHSAGLVHRDVKPGNILIT
Mvan_0024|M.vanbaalenii_PYR-1 EPLNSVIKRTGRLSLRHALDMLEQTGRALQVAHSAGLVHRDVKPGNILIT
MSMEG_0030|M.smegmatis_MC2_155 EPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHTAGLVHRDVKPGNILIT
MAB_0034c|M.abscessus_ATCC_199 EPLNSVLKRTGRLSLRHSLDMLEQTGRALQVAHAAGLVHRDVKPGNILIT
Mb0015c|M.bovis_AF2122/97 EPLNSVLKRTGRLSLRHALDMLEQTGRALQIAHAAGLVHRDVKPGNILIT
Rv0015c|M.tuberculosis_H37Rv EPLNSVLKRTGRLSLRHALDMLEQTGRALQIAHAAGLVHRDVKPGNILIT
MUL_0019|M.ulcerans_Agy99 EPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRDVKPGNILIT
MMAR_0017|M.marinum_M EPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRDVKPGNILIT
MAV_0019|M.avium_104 EPLNSVLKRTGRLSLRHALDMLEQTGRALQVAHAAGLVHRDVKPGNILIT
MLBr_00017|M.leprae_Br4923 EPLNSVLKRTGRLSLRHALDMLEQTGRALQIAHAAGLVHRDVKPGNILIT
TH_2631|M.thermoresistible__bu VTLRDIVHTDGPMEPKRAIEVIADACQALNFSHQHGIIHRDVKPANIMIS
.*..::: * :. ::::::: :: :**:.:* *::******.**:*:
Mflv_0811|M.gilvum_PYR-GCK PTGQVKLTDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGHDATAASD
Mvan_0024|M.vanbaalenii_PYR-1 PTGQVKLTDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGHDATAASD
MSMEG_0030|M.smegmatis_MC2_155 PTGQVKLTDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGHDATAASD
MAB_0034c|M.abscessus_ATCC_199 PTGQVKLTDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGHDATPASD
Mb0015c|M.bovis_AF2122/97 PTGQVKITDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGHDASPASD
Rv0015c|M.tuberculosis_H37Rv PTGQVKITDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGHDASPASD
MUL_0019|M.ulcerans_Agy99 PTGQVKITDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGQDASPSSD
MMAR_0017|M.marinum_M PTGQVKITDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGQDASPSSD
MAV_0019|M.avium_104 PTGQVKITDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGHDATPASD
MLBr_00017|M.leprae_Br4923 PTGQVKITDFGIAKAVDAAP-VTQTGMVMGTAQYIAPEQALGHDATPASD
TH_2631|M.thermoresistible__bu KTGAVKVMDFGIARAIADSNSVTQTAAVIGTAQYLSPEQARGETVDARSD
** **: *****:*: : ****. *:*****::**** *. . . **
Mflv_0811|M.gilvum_PYR-GCK VYSLGVVGYEAVSGKRPFTGDGALTVAMKHIKETPPPLP---ADLPPNVR
Mvan_0024|M.vanbaalenii_PYR-1 VYSLGVVGYEAVSGKRPFIGDGALTVAMKHIKETPPPLP---ADLPPNVR
MSMEG_0030|M.smegmatis_MC2_155 VYSLGVVGYESVSGKRPFTGDGALTVAMKHIKETPPPLP---ADLPPNVR
MAB_0034c|M.abscessus_ATCC_199 VYSLGVVGYEAVSGKRPFSGDGALTVAMKHIKETPSPLP---ADLPPNVR
Mb0015c|M.bovis_AF2122/97 VYSLGVVGYEAVSGKRPFAGDGALTVAMKHIKEPPPPLP---PDLPPNVR
Rv0015c|M.tuberculosis_H37Rv VYSLGVVGYEAVSGKRPFAGDGALTVAMKHIKEPPPPLP---PDLPPNVR
MUL_0019|M.ulcerans_Agy99 VYSLGVVGYEAVSGKRPFTGDGALTVAMKHIKEPPPPLP---AELPPNVR
MMAR_0017|M.marinum_M VYSLGVVGYEAVSGKRPFTGDGALTVAMKHIKEPPPPLP---AELPPNVR
MAV_0019|M.avium_104 VYSLGVVGYEVVSGKRPFSGDGALTVAMKHIKEPPPPLP---AELPPNVR
MLBr_00017|M.leprae_Br4923 VYSLGVIGYEVVSGKRPFTGDGALTVAMKHIKEPPPPLP---ADLPPNVR
TH_2631|M.thermoresistible__bu VYSLGCVLYEILTGEPPFVGDSPVAVAYQHVREDPVPPSRRNPAVSPELD
***** : ** ::*: ** **..::** :*::* * * . . :.*::
Mflv_0811|M.gilvum_PYR-GCK ELIEITLVKNPGMRYKSGGPFADAVAAVRAGRRPPRPNAAPSIGRAAPAA
Mvan_0024|M.vanbaalenii_PYR-1 ELIEITLVKNPGMRYKSGGPFADAVAAVRAGRRPPRPNAAPSIGRAAPTA
MSMEG_0030|M.smegmatis_MC2_155 ELIEITLVKNPGMRYKSGGPFADAVAAVRAGRRPPRPNQAPTLGRAAPAA
MAB_0034c|M.abscessus_ATCC_199 ELIEITLVKNPQNRYTSGGPFADAVAAVRAGRRPPRPNQAPTISRATPAA
Mb0015c|M.bovis_AF2122/97 ELIEITLVKNPAMRYRSGGPFADAVAAVRAGRRPPRPSQTPPPGRAAPAA
Rv0015c|M.tuberculosis_H37Rv ELIEITLVKNPAMRYRSGGPFADAVAAVRAGRRPPRPSQTPPPGRAAPAA
MUL_0019|M.ulcerans_Agy99 ELIEITLVKNPGLRYRSGGPFADAVAAVRAGRRPPRPSQSPPAGRAAPAA
MMAR_0017|M.marinum_M ELIEITLVKNPGLRYRSGGPFADAVAAVRAGRRPPRPSQSPPAGRAAPAA
MAV_0019|M.avium_104 ELIEITLVKNPAMRYRSGGPFADAVAAVRAGRRPPRPSQSPPPGRASPAA
MLBr_00017|M.leprae_Br4923 ELIEITLVKNPGMRYPSGGLFAEAVAAVRAGHRPPRPNQTPSSGRASPTT
TH_2631|M.thermoresistible__bu AVVLKALAKNPDNRYQTAAEMRADLVRVHSGEQPEAPKVLTDAERTSLLA
:: :*.*** ** :.. : :. *::*.:* *. . *:: :
Mflv_0811|M.gilvum_PYR-GCK VPPAIQNRPPAEATGRAPAQTSRTRTTGSHHRSAPPARRTFSSGQRALLW
Mvan_0024|M.vanbaalenii_PYR-1 VPPAIASRPAADPTGRAPAPTSRTRATGSHHRSAPPPRGGFSSGQRALLW
MSMEG_0030|M.smegmatis_MC2_155 VPSAAQARASADLTGRAPVTAARARPTATAAHRTPPPRRTFSSGQRALLW
MAB_0034c|M.abscessus_ATCC_199 IPPATQVRPAVGAPSRVAPPTSRTRQPTGSHRPPPPRRGTFSPGQKALMW
Mb0015c|M.bovis_AF2122/97 IPSGTTARVAANSAGRTAASR-RSRPATGGH-RPP--RRTFSSGQRALLW
Rv0015c|M.tuberculosis_H37Rv IPSGTTARVAANSAGRTAASR-RSRPATGGH-RPP--RRTFSSGQRALLW
MUL_0019|M.ulcerans_Agy99 IPSSAPARIAPAPTTRTTTPR-RTRPATGGH-RPPPSRRTFSSGQRALLW
MMAR_0017|M.marinum_M IPSSAPARIAPAPTTRTTTPR-RTRPATGGH-RPPPARRTFSSGQRALLW
MAV_0019|M.avium_104 IPSSPTTRAAAVSSGRTAAPR-RTRPATGGH-RPPPARRTFSSGQRALLW
MLBr_00017|M.leprae_Br4923 IPSSTQARAAVACGTKTPAPR-RSRPSTSGN-RPPPARNTFSSGQRALLW
TH_2631|M.thermoresistible__bu SGPAHDRTQPTDAVIDPVIPRYTGSDRGGSVGRWVAAVAVLAVLTVVVTV
.. . :: .:
Mflv_0811|M.gilvum_PYR-GCK AAGVLG------------------ALAIVIAILIVLNHQDQQNSPNRTTN
Mvan_0024|M.vanbaalenii_PYR-1 AAGVLG------------------ALAIVIAILIVLNAQDRKDQPPPTT-
MSMEG_0030|M.smegmatis_MC2_155 AAGVLG------------------ALAIVIAILIVLNAQDRKDREQSPPP
MAB_0034c|M.abscessus_ATCC_199 AAAVLG------------------IFAIIIAALILLQASEKKNQTPSIPA
Mb0015c|M.bovis_AF2122/97 AAGVLG------------------ALAIIIAVLLVIKAPGDNSPQQAPTP
Rv0015c|M.tuberculosis_H37Rv AAGVLG------------------ALAIIIAVLLVIKAPGDNSPQQAPTP
MUL_0019|M.ulcerans_Agy99 AAGVLG------------------ALAIIIAVLIVINSRAD-SQQQSPPP
MMAR_0017|M.marinum_M AAGVLG------------------ALAIIIAVLIVINSRAD-SQQQPPPP
MAV_0019|M.avium_104 AAGVLG------------------ALAIIIAVLIVINSRAD-QQQQ--PP
MLBr_00017|M.leprae_Br4923 AAGMLG------------------ALAIIIAVLIVINSYAGNEQHQPPTP
TH_2631|M.thermoresistible__bu AINMIGGNPRNVQVPDVSGQASADAIAALQNRGFKTRTQQQPDNSVPPDR
* ::* :* : : . .
Mflv_0811|M.gilvum_PYR-GCK TVTETP----GSPPAAPGEGPAETPGAAAPVKPD-EP------GQNRVSG
Mvan_0024|M.vanbaalenii_PYR-1 -VTETP----TSQGAPP-----ETPGAAAPAFGDHEP------DRYWVLR
MSMEG_0030|M.smegmatis_MC2_155 TVTDTV----TETTPYE-----ETPAAMP--------------DLMIMLR
MAB_0034c|M.abscessus_ATCC_199 TVSGSE----TPSSPGS-----ASPGALAP--------------ANIVAL
Mb0015c|M.bovis_AF2122/97 TVTTTG----NPPASN------TGGTDASP--RLNWT----ERGETRHSG
Rv0015c|M.tuberculosis_H37Rv TVTTTG----NPPASN------TGGTDASP--RLNWT----ERGETRHSG
MUL_0019|M.ulcerans_Agy99 TVTQTTTAHQTPTGQGPRLNWTDGQSIGNPGLQSNQPDLVDDAGRNRPIG
MMAR_0017|M.marinum_M TVTQTTTAHQTPTGQGPRLNWTEGKAIGNPGLQSNQPDLVDDAGRNRPIG
MAV_0019|M.avium_104 TVTDTG----TPPASAPPPTKTPSGSGAHPGLRLDWP----DDGTIGASG
MLBr_00017|M.leprae_Br4923 TVTDTG----TPPATKT----LSGFPAAYCEYRVNWT----NHKEISNSG
TH_2631|M.thermoresistible__bu VISTDPPADTSVSAGDEITVNVSTGPQQREIPDVSSLTYAEAVRRLSAAG
::
Mflv_0811|M.gilvum_PYR-GCK PVD------TASPSVMLHVSEQILR-------------------------
Mvan_0024|M.vanbaalenii_PYR-1 PVD------NLSPLTTRHVSEQNLR-------------------------
MSMEG_0030|M.smegmatis_MC2_155 AAQ------PEPPP-----SEQIQR-------------------------
MAB_0034c|M.abscessus_ATCC_199 LPD------AEPDPIPGTKSP-----------------------------
Mb0015c|M.bovis_AF2122/97 LQS-----WVVPPTPHSRASLARYEIAQ----------------------
Rv0015c|M.tuberculosis_H37Rv LQS-----WVVPPTPHSRASLARYEIAQ----------------------
MUL_0019|M.ulcerans_Agy99 NRD-SATSWNVTVTPQHRAALARFEMQR----------------------
MMAR_0017|M.marinum_M NRG-SATSWNVTVTPQHLAALARFEMQR----------------------
MAV_0019|M.avium_104 FRDGPARHWTSQ--------------------------------------
MLBr_00017|M.leprae_Br4923 LPK-----QAARAQLAGAIDISPVAGQT----------------------
TH_2631|M.thermoresistible__bu FEKFRQSESPSTPELRGKVVATNPPANQTAAITNEITVVVGTGPATREVP
Mflv_0811|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0024|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0030|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0034c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0015c|M.bovis_AF2122/97 --------------------------------------------------
Rv0015c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_0019|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0017|M.marinum_M --------------------------------------------------
MAV_0019|M.avium_104 --------------------------------------------------
MLBr_00017|M.leprae_Br4923 --------------------------------------------------
TH_2631|M.thermoresistible__bu DITGQTVELAERNLSVYGFTKFTQSAVDSTAPAGQVVGTNPPAGTTVSLD
Mflv_0811|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0024|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0030|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0034c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0015c|M.bovis_AF2122/97 --------------------------------------------------
Rv0015c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_0019|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0017|M.marinum_M --------------------------------------------------
MAV_0019|M.avium_104 --------------------------------------------------
MLBr_00017|M.leprae_Br4923 --------------------------------------------------
TH_2631|M.thermoresistible__bu SVIELEVSKGNQFVMPDLIGMFWTDAEPRLRALGWTGVLDKGADIPNSGQ
Mflv_0811|M.gilvum_PYR-GCK ----------------------------
Mvan_0024|M.vanbaalenii_PYR-1 ----------------------------
MSMEG_0030|M.smegmatis_MC2_155 ----------------------------
MAB_0034c|M.abscessus_ATCC_199 ----------------------------
Mb0015c|M.bovis_AF2122/97 ----------------------------
Rv0015c|M.tuberculosis_H37Rv ----------------------------
MUL_0019|M.ulcerans_Agy99 ----------------------------
MMAR_0017|M.marinum_M ----------------------------
MAV_0019|M.avium_104 ----------------------------
MLBr_00017|M.leprae_Br4923 ----------------------------
TH_2631|M.thermoresistible__bu RANAVVRQNPSPGAGVNYDARITLNFAS