For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPGPAATARRRRRWGRPGIGLRLLAAQVIVLVAGAATTAVVAAVVGPPLFREHLHLAGVPTDSPEEIHAE EAYGYATVMSIGGALAVSALAALAVSFYVSRRLQRSITEVASAATDVAQGNYEIRVSPPGLGDDFDALST AFNQMASRLQAVESTRQRLFGDLAHEIRTPVAVLEAYIEALEDGVRTLTPQTAAMLRDQTRRLVRFSDDV AALAKAEESSVSMSYATIDVDRLTRQYVAAAQKRYDSKGVALRVRLQTTLPPLWADEQRLSQVLGNLLEN ALRHTPSGGSVELTGVRDGDRLRIAVADTGEGIAPEHLPRLFERFYRADAARDREHGGAGLGLAIAKALV EAHGGSISVASAGLGAGATFTVDLPVAQVRADDVPVTGASART*
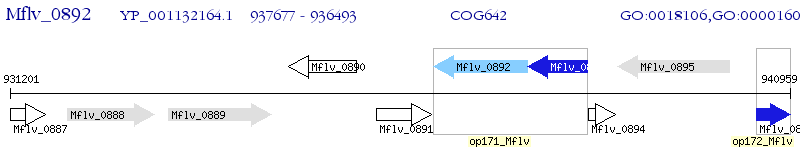
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0892 | - | - | 100% (394) | integral membrane sensor signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_2837 | - | 0.0 | 86.51% (393) | integral membrane sensor signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_3888 | - | e-157 | 77.33% (375) | integral membrane sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0781 | phoR | 2e-34 | 31.35% (303) | two component system response sensor kinase membrane |
| M. tuberculosis H37Rv | Rv0758 | phoR | 2e-34 | 30.45% (312) | two component system response sensor kinase membrane |
| M. leprae Br4923 | MLBr_02440 | - | 3e-29 | 34.50% (229) | putative two-component system sensor histidine kinase |
| M. abscessus ATCC 19977 | MAB_3340 | - | 1e-107 | 57.50% (360) | sensor histidine kinase |
| M. marinum M | MMAR_4941 | phoR | 2e-34 | 30.84% (334) | two-component system response phosphate sensor kinase, PhoR |
| M. avium 104 | MAV_0703 | - | 3e-40 | 33.13% (326) | sensor histidine kinase PhoR |
| M. smegmatis MC2 155 | MSMEG_5870 | - | 6e-38 | 32.41% (324) | sensor histidine kinase PhoR |
| M. thermoresistible (build 8) | TH_2452 | - | 7e-34 | 31.19% (295) | PUTATIVE putative sensor histidine kinase PhoR |
| M. ulcerans Agy99 | MUL_0464 | phoR | 9e-35 | 30.84% (334) | two component system response phosphate sensor kinase, PhoR |
| M. vanbaalenii PYR-1 | Mvan_1833 | - | 1e-159 | 75.85% (381) | integral membrane sensor signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0892|M.gilvum_PYR-GCK -------------------------------------MTPGPAATARRRR
Mvan_1833|M.vanbaalenii_PYR-1 -------------------------------------MARTEEELRPRRR
MAB_3340|M.abscessus_ATCC_1997 -------------------------------------MTR----------
MMAR_4941|M.marinum_M -----------MAERLPRAVPLRVGLVAATLALVLCGLLASGVAVTSILR
MUL_0464|M.ulcerans_Agy99 -----------MAERLPRAVQLRVGLVAATLALVLCGLLASGVAVTSILR
MAV_0703|M.avium_104 ----------MVVTRPRRGLPLRVGLVAATLALVACGLAVSGIAVTSILR
MSMEG_5870|M.smegmatis_MC2_155 ------------MNRGRRGIPLRVGLVAAALVLVACGLLASGIAVTSIMR
Mb0781|M.bovis_AF2122/97 -----------MARHLRGRLPLRVRLVAATLILVATGLVASGIAVTSMLQ
Rv0758|M.tuberculosis_H37Rv -----------MARHLRGRLPLRVRLVAATLILVATGLVASGIAVTSMLQ
TH_2452|M.thermoresistible__bu MTSEPPSPSGRPRWRRLLPSAARTRIIGWVLLLVMVALGVVTLTTWRLMV
MLBr_02440|M.leprae_Br4923 -------------------------------MFSALLLAGVLSGLAFVVG
:
Mflv_0892|M.gilvum_PYR-GCK RWGRPGIGLRLLAAQVIVLVAGAA-----------------TTAVVAAVV
Mvan_1833|M.vanbaalenii_PYR-1 MLRRPGIGLRLLAAQAIVLVAGAV-----------------TTSVVAAIV
MAB_3340|M.abscessus_ATCC_1997 --PRYGIGLRLLVVNSLVVLAGIA-----------------TTSVVAAIV
MMAR_4941|M.marinum_M HSLVSRIDSTLLEASRTWAQASWR-----------------HSMPSVEGP
MUL_0464|M.ulcerans_Agy99 HSLVSRIDSTLLEASRTWAQASWR-----------------HSMPSVEGP
MAV_0703|M.avium_104 HSLVSRIDSTLLDASRGWAQAPRR-----------------QSSSAYEGP
MSMEG_5870|M.smegmatis_MC2_155 HTEVSRIDQTLLDASRSWAQAPRR-----------------APTTPLEGP
Mb0781|M.bovis_AF2122/97 HRLTSRIDRVLLEEAQIWAQITLP-----------------LAPDPYPIH
Rv0758|M.tuberculosis_H37Rv HRLTSRIDRVLLEEAQIWAQITLP-----------------LAPDPYPGH
TH_2452|M.thermoresistible__bu SSIDARMDEALHIEVDEFEELTASGLNPRTGAPFTGVAEVIREAIAYNLA
MLBr_02440|M.leprae_Br4923 IVAGIRLSPRLIERRQRLANEWAG---------------------ITVLQ
:. *
Mflv_0892|M.gilvum_PYR-GCK GPP----------LFREHLHLAGVPTDSPEEIHAEEAYG--------YAT
Mvan_1833|M.vanbaalenii_PYR-1 GPP----------LFHDHLHQAGISAGSPEEMHAEEAYT--------YAT
MAB_3340|M.abscessus_ATCC_1997 GPP----------MFRRLMDQQ-VSPGAGNDHPYERAFR--------DAT
MMAR_4941|M.marinum_M DPAR-----PPSKFYVRSISPDGRAMTAINDRNAEPALPADNDVGSDPTT
MUL_0464|M.ulcerans_Agy99 DPAR-----PPSKFYVRSISPDGRAMTAINDRNAEPALPADNDVGSDPTT
MAV_0703|M.avium_104 DPGR-----PPSKFYVRGVSTDGTPFTAINDRNAEPALPADNDVGPNPTT
MSMEG_5870|M.smegmatis_MC2_155 NPAR-----PPSNFYVRGFDEDGRTWIAVNDREAEPALPENNDVGPVPVT
Mb0781|M.bovis_AF2122/97 NPDR-----PPSRFYVRVISPDGQSYTALNDNTAIPAVPANNDVGRHPTT
Rv0758|M.tuberculosis_H37Rv NPDR-----PPSRFYVRVISPDGQSYTALNDNTAIPAVPANNDVGRHPTT
TH_2452|M.thermoresistible__bu RPNEKFLGYVDGEYHTQSRQEPGFPEVLAGDDAFTERVATVTEPVEGSYD
MLBr_02440|M.leprae_Br4923 MLQR----------IIALMPLGAAVVDTYRDVVYLNEQAK----------
:
Mflv_0892|M.gilvum_PYR-GCK VMSIGG----------------------------------------ALAV
Mvan_1833|M.vanbaalenii_PYR-1 VISIGG----------------------------------------ALAV
MAB_3340|M.abscessus_ATCC_1997 AMSIGF----------------------------------------AVAV
MMAR_4941|M.marinum_M LPSVNGSNIEWRAVSVHGPQGGLTTVAIDLSDVQ-HTVRSLVWLQIGIGA
MUL_0464|M.ulcerans_Agy99 LPSVNGSNIEWRAVSVHGPQGGLTTVAIDLSDVQ-HTVRSLVWLQIGIGA
MAV_0703|M.avium_104 LPSVNGSDIQWRAVSVRGPHG-LTTVAIDLSDVQ-HTVSSLVWLQIGIGV
MSMEG_5870|M.smegmatis_MC2_155 VGSIDKSNVEWRAMTVRGPNGELTTVAIDLSEVQ-SSVRALVWSQVGIGA
Mb0781|M.bovis_AF2122/97 LPSIGGSKTLWRAVSVRASDGYLTTVAIDLADVR-STVRSLVLLQVGIGS
Rv0758|M.tuberculosis_H37Rv LPSIGGSKTLWRAVSVRASDGYLTTVAIDLADVR-STVRSLVLLQVGIGS
TH_2452|M.thermoresistible__bu HPEVGEVRYLAIPVTMAGDPAQGVIVAAYLGDAERAAADDAARLMLLVGA
MLBr_02440|M.leprae_Br4923 ----------------------------------------------ELGL
:.
Mflv_0892|M.gilvum_PYR-GCK SALAALAVS-FYVSRRLQR----SITEVASAATDVAQGNYEIRVSPPGLG
Mvan_1833|M.vanbaalenii_PYR-1 SALAALAVS-WYISRRLQR----SVTEVAEAASSVAEGHYDVRVSSPQLG
MAB_3340|M.abscessus_ATCC_1997 SALAALTLS-WYLSHRVHR----SATALSQAASAVADGNYDIRVPPPRLG
MMAR_4941|M.marinum_M GVLVVVGLAGFAVVQRSLR----PLSEVEQTAAAIASGQLDRRVPERDPR
MUL_0464|M.ulcerans_Agy99 GVLVVVGLAGFAVVQRSLR----PLSEVEQTAAAIASGQLDRRVPERDPR
MAV_0703|M.avium_104 AVLAVVGIASFAVVQRSLR----PLVEVEQTAAAIAAGQLDRRVPERDPR
MSMEG_5870|M.smegmatis_MC2_155 AVLLVLGVVGYVVVHRSLR----PLVEVERTAAAIAAGQLDRRVPERDPR
Mb0781|M.bovis_AF2122/97 AVLVVLGVAGYAVVRRSLR----PLAEFEQTAAAIGAGQLDRRVPQWHPR
Rv0758|M.tuberculosis_H37Rv AVLVVPGVAGYAVVRRSLR----PLAEFEQTAAAIGAGQLDRRVPQWHPR
TH_2452|M.thermoresistible__bu ATLFGAIGAAWLVAGRILR----PLRDVADTARTITDTDLSRRIPSRGEG
MLBr_02440|M.leprae_Br4923 VRDRQLDDQAWRAAQQALGGDDVEFDLLPGKRPAAGRSGLSVHGHARLLS
: : . . :
Mflv_0892|M.gilvum_PYR-GCK DDFDALSTAFNQMASRLQAVESTR--------------QRLFGDLAHEIR
Mvan_1833|M.vanbaalenii_PYR-1 DDFDALADAFNQMASRLQTVESTR--------------QRLFSDLAHEIR
MAB_3340|M.abscessus_ATCC_1997 KEFDAVAAAFNKMAQRLGAVDEAR--------------RQMLADLAHEIR
MMAR_4941|M.marinum_M TEVGQLSLAMNGMLSQIQVALAASEDSAEKARSSEERMRRFITDASHELR
MUL_0464|M.ulcerans_Agy99 TEVGQLSLAMNGMLSQIQVALAASEDSAEKARSSEERMRRFITDASHELR
MAV_0703|M.avium_104 TEVGRLSLALNGMLAQIQQALASSESSAEKARGSEDRMRRFITDASHELR
MSMEG_5870|M.smegmatis_MC2_155 TEVGRLSSALNGMLAQIQRAVAASEASAEQARSSEDRMRRFITDASHELR
Mb0781|M.bovis_AF2122/97 TEVGRLSLALNGMLAQIQRAVASAESSAEKARDSEDRMRQFITDASHELR
Rv0758|M.tuberculosis_H37Rv TEVGRLSLALNGMLAQIQRAVASAESSAEKARDSEDRMRQFITDASHELR
TH_2452|M.thermoresistible__bu DEMDSLVRTINGMLDRIEAGVSAQ--------------RRFIDDAGHELR
MLBr_02440|M.leprae_Br4923 EKDRRFAVVFVHDQSDYVRMEAAR--------------RDFVANVSHELK
. . .: : : :. : .**::
Mflv_0892|M.gilvum_PYR-GCK TPVAVLEAYIEALED--GVRTLTPQTAAMLRDQTRRLVRFSDDVAALAKA
Mvan_1833|M.vanbaalenii_PYR-1 TPVAVLEAYIEALED--GVRTLTPQTAAMLRDQTRRLVRFSDDVAALAKA
MAB_3340|M.abscessus_ATCC_1997 TPVAVLEAYMEAVED--GVQQLDQQTVATLRDQTRRLTRFSIDVNALSVA
MMAR_4941|M.marinum_M TPLTTIRGFAELYRQ--GAARDVAMLLSRIESEASRMGLLVDDLLTLARL
MUL_0464|M.ulcerans_Agy99 TPLTTIRGFAELYRQ--GAARDVAMLLSRIESEASRMGLLVDDLLTLARL
MAV_0703|M.avium_104 TPLTTIRGFAELYRQ--GAARDVAMLLSRIESEASRMGLLVDDLLLLARL
MSMEG_5870|M.smegmatis_MC2_155 TPLTTIRGFAELYRQ--GAARDIEMLMGRIESESRRMGLLVEDLLLLARL
Mb0781|M.bovis_AF2122/97 TPLTTIRGFAELYRQ--GAARDVGMLLSRIESEASRMGLLVDDLLLLARL
Rv0758|M.tuberculosis_H37Rv TPLTTIRGFAELYRQ--GAARDVGMLLSRIESEASRMGLLVDDLLLLARL
TH_2452|M.thermoresistible__bu TPITIVRGHLEVLDP--DDPDDVRSTVALVDDELVRMNRMVTDLLLLARS
MLBr_02440|M.leprae_Br4923 TPVGAMALLAEALLASADDAETVSRFAEKVLIEANRLGYMVAELIELSRL
**: : * . : : *: : :: *:
Mflv_0892|M.gilvum_PYR-GCK EESSVSMSYATIDVDRLTRQYVAAAQKRYDSKGVALRVRLQTTLPPLWAD
Mvan_1833|M.vanbaalenii_PYR-1 EESSASMACSFVDVPRLAHHCAAAAQERFDSKNVTLAVRVAEGLPPLWAD
MAB_3340|M.abscessus_ATCC_1997 ESRTTSIDARSVSPADLVTTAVAAFAPRYQTKGVILTSHADDGLPDVWAD
MMAR_4941|M.marinum_M DVQRP-LERNRVDLLVLAADAVHDARAIDRKRAITVEVLEGPGTPEVLGD
MUL_0464|M.ulcerans_Agy99 DVQRP-LERNRVDLLVLAADAVHDARAIDRKRAITVEVLEGPGTPEVLGD
MAV_0703|M.avium_104 DVQRP-LEHHRVDLLALASDAVHDAQAMDPKRTITLEVLDGPGTPEVFGD
MSMEG_5870|M.smegmatis_MC2_155 DAQRP-LERRRVDLLALATDAVHDAQSIAPKRRVKMEVFDGPGTPEVLGD
Mb0781|M.bovis_AF2122/97 DAHRP-LELCRVDLLALASDAAHDARAMDPKRRITLEVLDGPGTPEVLGD
Rv0758|M.tuberculosis_H37Rv DAHRP-LELCRVDLLALASDAAHDARAMDPKRRITLEVLDGPGTPEVLGD
TH_2452|M.thermoresistible__bu E-QPTFLTPEPTDIGALTRAIFDKVVQLGDRDYQLQSVAEVT----AVLD
MLBr_02440|M.leprae_Br4923 QGAERLPNVTDIDVDIIVSEAIARHKVAADTAAIEVRTDPPSGLR-VLGD
: . :. *
Mflv_0892|M.gilvum_PYR-GCK EQRLSQVLGNLLENALRHTPSGGSVELTGVRDGDRLRIAVADTGEGIAPE
Mvan_1833|M.vanbaalenii_PYR-1 EQRLAQVVGNLVDNALRHTPAGGRVEVSWEREGAELALRVADNGAGIAAE
MAB_3340|M.abscessus_ATCC_1997 PERMGQVLSNLLDNALRHTGPGGQVTVTATATGTGATIAVADTGDGVPAE
MMAR_4941|M.marinum_M EPRLRQVLSNLVGNALQHTPDSADVTVRVGTAGQNAVLEVADKGPGMPAE
MUL_0464|M.ulcerans_Agy99 EPRLRQVLSNLVGNALQHTPDSADVTVRVGTAGENAVLEVADKGPGMPAE
MAV_0703|M.avium_104 EPRIRQVLGNLIANALQHTPESADVTVRVGTDGDDAVLEVADRGPGMNEQ
MSMEG_5870|M.smegmatis_MC2_155 EARLRQVIGNLVANAVQHTPETAGITVRVGTDGDNAVLEVCDEGPGMSQE
Mb0781|M.bovis_AF2122/97 ESRLRQVLRNLVANAIQHTPESADVTVRVGTEGDDAILEVADDGPGMSQE
Rv0758|M.tuberculosis_H37Rv ESRLRQVLRNLVANAIQHTPESADVTVRVGTEGDDAILEVADDGPGMSQE
TH_2452|M.thermoresistible__bu AQRITQALIALADNACRYTRDGDWIAVGSGLYDGWLRFWVSDSGPGVSDA
MLBr_02440|M.leprae_Br4923 QTLLVTALANLVSNAIAYSPGGSLVSISRRRRGDNIEIAVTDRGIGIALE
: .: * ** :: : : . : * * * *:
Mflv_0892|M.gilvum_PYR-GCK HLPRLFERFYRADAARDREHGGAGLGLAIAKALVEAHGGSISVASAGLGA
Mvan_1833|M.vanbaalenii_PYR-1 HLPRLFERFYRADTARDREHGGAGIGLAITRALVEAHGGTISVASAGPGT
MAB_3340|M.abscessus_ATCC_1997 HLDRLFDRFYRADAARDRQHGGAGIGLSIAKALVEAHEGRIEAHSAGHAA
MMAR_4941|M.marinum_M DAARVFERFYRTDSSRARASGGTGLGLSIVHSLVKAHGGDVTLTTA-PGE
MUL_0464|M.ulcerans_Agy99 DAARVFERFYRTDSSRARASGGTGLGLSIVHSLVKAHGGDVTLTTA-PGE
MAV_0703|M.avium_104 DASRVFERFYRTDSSRARASGGTGLGLSIVESLVRAHGGTVGVTTA-PGQ
MSMEG_5870|M.smegmatis_MC2_155 DARRVFERFYRADTSRTRASGGTGLGLSIVDSLVYAHGGTVKVTTA-LGK
Mb0781|M.bovis_AF2122/97 DALRVFERFYRADSSRARASGGTGLGLSIVDSLVAAHGGAVTVTTA-LGE
Rv0758|M.tuberculosis_H37Rv DALRVFERFYRADSSRARASGGTGLGLSIVDSLVAAHGGAVTVTTA-LGE
TH_2452|M.thermoresistible__bu DRDRIFERFARGSAGGQRSDG-AGLGLSIVRAIAGAHGGHVDLDTV-VGR
MLBr_02440|M.leprae_Br4923 DQERVFERFFRGDKARSRATGGSGLGLAIVKHVAANHNGSIGVWSK-PGT
. *:*:** * . . * * :*:**:*. :. * * : : .
Mflv_0892|M.gilvum_PYR-GCK GATFTVDLPVAQVRADDVPVTGASART-----------------------
Mvan_1833|M.vanbaalenii_PYR-1 GATFTVTLPLAPARDELVALGVARPAAQ----------------------
MAB_3340|M.abscessus_ATCC_1997 GTTFTVSLPRPSDHPEKAP--GQRPG------------------------
MMAR_4941|M.marinum_M GCCFRVTLPRVSEAAVELTEPVS---------------------------
MUL_0464|M.ulcerans_Agy99 GCCFRVTLPRVSEAAVELTEPVS---------------------------
MAV_0703|M.avium_104 GCCFRVTLPRISDVPAVQAS------------------------------
MSMEG_5870|M.smegmatis_MC2_155 GSRFTVSLPRIADLPARL--------------------------------
Mb0781|M.bovis_AF2122/97 GCCFRVSLPRVSDVDQLSLTPVVPGPP-----------------------
Rv0758|M.tuberculosis_H37Rv GCCFRVSLPRVSDVDQLSLTPVVPGPP-----------------------
TH_2452|M.thermoresistible__bu GSTFTITIPAVLEAPWPAS-------------------------------
MLBr_02440|M.leprae_Br4923 GSTFTLSIPAAMPLYQDNDEQSGQPRGCDMWLNRPQREEEEFKSMTPAQA
* * : :*
Mflv_0892|M.gilvum_PYR-GCK -------------------------
Mvan_1833|M.vanbaalenii_PYR-1 -------------------------
MAB_3340|M.abscessus_ATCC_1997 -------------------------
MMAR_4941|M.marinum_M -------------------------
MUL_0464|M.ulcerans_Agy99 -------------------------
MAV_0703|M.avium_104 -------------------------
MSMEG_5870|M.smegmatis_MC2_155 -------------------------
Mb0781|M.bovis_AF2122/97 -------------------------
Rv0758|M.tuberculosis_H37Rv -------------------------
TH_2452|M.thermoresistible__bu -------------------------
MLBr_02440|M.leprae_Br4923 MMQSEVTRGNVNDKCPDCGGRGIAG