For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSLLCTSPARRYGMGFERIDDLLHRVTGDGSLHGVAATVVGRDGVLYEGASGSATPNSMFRNASMTKAVA TTGALQLVEQGRVDLDAPVSSIVPEFGELQVLEGFDDGAPVLRAPRTQATVRQLMTHTAGCGYHFLNENL YRYCTEFGFPDPFSGMKKSISAPLVHDPGTVWEYGVNTDWLGMVVEAVSGQTLGDYLTEHVYRPLGMTDS TFAPSDEQRARLLPVHSRAADGSLAPTDLDLPEAPQWDAAGHGSYGTVADYGRFVRAWLNDGELDGTRIL KAETVELALRDHLDGAPLPTKMEPTVPELAKAVELLPVPQGWGLGFHLYHVDLPGMRSAGSGDWSGLFNS FFWIDRTAGIGGVIATQLLPFFDDTMVETILAFEAAVYASL*
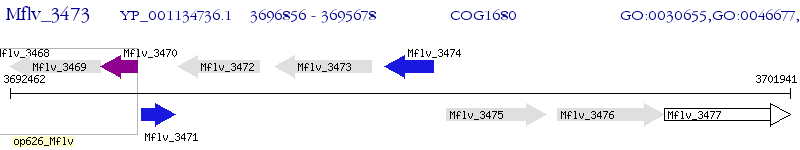
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3473 | - | - | 100% (392) | beta-lactamase |
| M. gilvum PYR-GCK | Mflv_3749 | - | 2e-30 | 30.63% (333) | beta-lactamase |
| M. gilvum PYR-GCK | Mflv_0495 | - | 4e-17 | 29.83% (238) | beta-lactamase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1402c | - | 7e-26 | 29.34% (317) | hypothetical protein Mb1402c |
| M. tuberculosis H37Rv | Rv1367c | - | 7e-26 | 29.34% (317) | hypothetical protein Rv1367c |
| M. leprae Br4923 | MLBr_00119 | lipE | 1e-07 | 24.00% (250) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_2833 | - | 1e-29 | 28.89% (360) | beta-lactamase |
| M. marinum M | MMAR_2187 | - | 1e-25 | 28.53% (319) | beta-lactamase |
| M. avium 104 | MAV_3395 | - | 4e-29 | 30.45% (335) | beta-lactamase |
| M. smegmatis MC2 155 | MSMEG_3040 | - | 9e-28 | 29.73% (333) | beta-lactamase |
| M. thermoresistible (build 8) | TH_4016 | - | 7e-21 | 35.63% (174) | PUTATIVE beta-lactamase |
| M. ulcerans Agy99 | MUL_1772 | - | 6e-26 | 28.53% (319) | beta-lactamase |
| M. vanbaalenii PYR-1 | Mvan_1072 | - | 0.0 | 82.28% (378) | beta-lactamase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3473|M.gilvum_PYR-GCK --------------------MSSLLCTS-----PARRYGMGFERIDDLLH
Mvan_1072|M.vanbaalenii_PYR-1 -----------MVGSLISVGMPDAACDDGPGVPEAEELYMGFERIDDLLS
Mb1402c|M.bovis_AF2122/97 --------------------------------------------------
Rv1367c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2187|M.marinum_M ---------------------------------------MNLDANQADIR
MUL_1772|M.ulcerans_Agy99 MGVARPRWRGPRAAWFAGARKPSDGRRLTRRPPAANLNPVNLDANQADIR
MAV_3395|M.avium_104 ---------------------------------------MTLDANSVSIR
MSMEG_3040|M.smegmatis_MC2_155 ---------------------------------------MNLDGNQTSIR
MAB_2833|M.abscessus_ATCC_1997 ----------------------------------MAEPDSTFDAALAPIR
TH_4016|M.thermoresistible__bu --------------------------------------------VIGQVD
MLBr_00119|M.leprae_Br4923 ----------MTSDGKIRVPTDLDAVTAIGDEDHSDIDSAAVERIWQATR
Mflv_3473|M.gilvum_PYR-GCK RVTGDGSLHGVAATVVGRDGVLYEGASG------------------SATP
Mvan_1072|M.vanbaalenii_PYR-1 EVTADGSLHGVAATVVGRDGVLYEGAAG------------------DAKH
Mb1402c|M.bovis_AF2122/97 -------------MVWQREKLLQVNEIGYRDIDA----------GVPMQR
Rv1367c|M.tuberculosis_H37Rv -------------MVWQREKLLQVNEIGYRDIDA----------GVPMQR
MMAR_2187|M.marinum_M EVCDAGLLSGAVTVVWQHGEVLQVNEIGYRDVEA----------GLPMQR
MUL_1772|M.ulcerans_Agy99 EVCDAGLLSGAVTVVWQHGEVLQVNEIGYRDVEA----------GLPMQR
MAV_3395|M.avium_104 EVCDAGLLAGAVTMVWQRGELLQVNEIGYRDVEA----------GLPMRR
MSMEG_3040|M.smegmatis_MC2_155 EAIDAGLLAGAVTLVWQAGKVLQVNTLGHRDVEA----------RLPMQR
MAB_2833|M.abscessus_ATCC_1997 AAIDDRILAGAVTLVWQGGQLRHLGATGYRDIDA----------GLSMAE
TH_4016|M.thermoresistible__bu DAVAVRRLPGAVVQIGHAGEVVFRQAFGSRKLAGEPGPTGEPTPAEPMTE
MLBr_00119|M.leprae_Br4923 HWYQAGMHPAIQLCIRQHGRVVLNRAIGHGWGNAPTDP--HDAEKIPVTT
: : *
Mflv_3473|M.gilvum_PYR-GCK NSMFRNASMTKAVA-TTGALQLVEQGRVDLDAPVSSIVPEFGELQVLEGF
Mvan_1072|M.vanbaalenii_PYR-1 DTMFRNASMTKAVA-TTAALQLVEQGRVDLDATVASLVPEFGELQVIDGF
Mb1402c|M.bovis_AF2122/97 DTLFRIASMTKPVT-VAAAMSLVDEGKLALRDPITRWAPELCKVAVLDDA
Rv1367c|M.tuberculosis_H37Rv DTLFRIASMTKPVT-VAAAMSLVDEGKLALRDPITRWAPELCKVAVLDDA
MMAR_2187|M.marinum_M DTLFRIASMTKPVT-VAAAMSMVDEGKMALRDPITRWAPELRDIRVLDDP
MUL_1772|M.ulcerans_Agy99 DTLFRIASMTKPVT-VAAAMSMVDEGKIALRDPITRWAPELRDIRVLDDP
MAV_3395|M.avium_104 DTLFRIASMTKPVT-VAAVMSLVDAGKLSLKDPVTRWAPELAGVRVLDDP
MSMEG_3040|M.smegmatis_MC2_155 DTIFRIASMTKPVT-VAAAMALAEEGKFALADPVTAWLPELSNMRVLREP
MAB_2833|M.abscessus_ATCC_1997 NTIFRIASMTKPII-SAATMNLVDDGTIRLSDPITTWLPEFSDMRVLKNP
TH_4016|M.thermoresistible__bu DTMFDLASLTKSLATTVAVLQLAEQGRLRVDDPVQSHLPEFN--------
MLBr_00119|M.leprae_Br4923 DTPFCAYSAAKAIT-ATVVHMLVERGHFSLDDRVCEYLPTYTSYG-----
:: * * :*.: . . :.: * . : : *
Mflv_3473|M.gilvum_PYR-GCK DDGAPVLRAPRTQATVRQLMTHTAGCGYHFLN-ENLYRYCTEFGFP---D
Mvan_1072|M.vanbaalenii_PYR-1 QGSEPVLRAPKTQATVRQLMNHTAGCGYHFLN-GELLAYCEGQNFP---N
Mb1402c|M.bovis_AF2122/97 AGPLDRTHPARRAILIEDLLTHTSGLAYGFSVSGPISRAYQRLPFG--QG
Rv1367c|M.tuberculosis_H37Rv AGPLDRTHPARRAILIEDLLTHTSGLAYGFSVSGPISRAYQRLPFG--QG
MMAR_2187|M.marinum_M HGPLDRTHPTRRPILIEDLLTHTSGLAYSFSVSGDISRAYMRLPFG--HG
MUL_1772|M.ulcerans_Agy99 HGPLDRTHPTRRPILIEDLLTHTSGLAYSFSVSGDISRAYMRLPFG--HG
MAV_3395|M.avium_104 HGPLDRTHPLDRAILIEDLLTHTSGLAYGFSVSGPISKAYMRLPFQ--HG
MSMEG_3040|M.smegmatis_MC2_155 RGPLDQTVPARRPITFDDLMTHRSGMAYVFSVLGPLASAYGKISQR--QD
MAB_2833|M.abscessus_ATCC_1997 EGPLDDTFRAPRLITVEDLLTHRSGLTYDFISTGPIAKAYHPLHTAAFRE
TH_4016|M.thermoresistible__bu ----PQHDPRRARVTLRMLLTHTSGIAGDLSLDGPWG-----LDRP---D
MLBr_00119|M.leprae_Br4923 ----------KHRTTIRHVLTHSAGVPFPTGPRPDVTRADDHAYAQ----
. ::.* :*
Mflv_3473|M.gilvum_PYR-GCK PFSGMKKSISAPLVHDPGTVWEYG-VNTDWLG-MVVEAVSGQTLGDYLTE
Mvan_1072|M.vanbaalenii_PYR-1 PLEGVKKSLSAPLVHDPGTVWEYG-VSTDWLG-MVVEAVSGQTLADYLAE
Mb1402c|M.bovis_AF2122/97 PDVWLAALATLPLVHQPGDRVTYS-HAIDVLG-VIVSRIEDAPLYQVIDE
Rv1367c|M.tuberculosis_H37Rv PDVWLAALATLPLVHQPGDRVTYS-HAIDVLG-VIVSRIEDAPLYQIIDE
MMAR_2187|M.marinum_M SDAWLAELAALPLVHQPGERVTYS-HAIDLLG-VIMSRIDDKPFYQVLDE
MUL_1772|M.ulcerans_Agy99 SDAWLAELAALPLVHQPGERVTYS-HAIDLLG-VIMSRIDDKPFYQVLDE
MAV_3395|M.avium_104 PDAWLAELAKLPLVHQPGEKVTYS-HSIDLLG-VIAARIEDKPFHQVLDE
MSMEG_3040|M.smegmatis_MC2_155 QDRWLAEVAALPLAHQPGERLTYS-HSTDVLG-IALSRIEGKPLSQVLSE
MAB_2833|M.abscessus_ATCC_1997 PDEWLAAIAALPLVYPPGERFHYS-HSTDVLG-LLIARAAGLPLNTLLRQ
TH_4016|M.thermoresistible__bu KAEGVRRALGAWVVHEPGRLFHYSDIGFILLG-AIVEKLTGEPLDVYVTE
MLBr_00119|M.leprae_Br4923 -----QKLSELRPFYRPGLVHIYHALTWGPLMREIIWTATGKEIRDILAA
: ** * * . : :
Mflv_3473|M.gilvum_PYR-GCK HVYRPLGMTDSTFAPSDEQR------------------------------
Mvan_1072|M.vanbaalenii_PYR-1 HVYGPLGMTDSTFDLTDEQR------------------------------
Mb1402c|M.bovis_AF2122/97 RVLGPAGMTDTGFYVSADAQ------------------------------
Rv1367c|M.tuberculosis_H37Rv RVLGPAGMTDTGFYVSADAQ------------------------------
MMAR_2187|M.marinum_M RILGPAGMTDTGFFVSTQAQ------------------------------
MUL_1772|M.ulcerans_Agy99 RILGPAGMTDTGFFVSTQAQ------------------------------
MAV_3395|M.avium_104 RVLGPAGMTDTGFFVSPQAR------------------------------
MSMEG_3040|M.smegmatis_MC2_155 RVFEPLGMTDTGFSIGAAGR------------------------------
MAB_2833|M.abscessus_ATCC_1997 RILDPLGMNDTDFFVPEHKA------------------------------
TH_4016|M.thermoresistible__bu NVFAPLGMADTRYLPVSKACGPIRMVGNAVVEDESAEPTPCGEDRWSTSL
MLBr_00119|M.leprae_Br4923 EILDPLGFRWTNFGVAEEDVPLVALSHATG--------------------
.: * *: : :
Mflv_3473|M.gilvum_PYR-GCK -ARLLPVHSRAAD-------GSLAPTDLDLPEAPQ-----WDAAGHGSYG
Mvan_1072|M.vanbaalenii_PYR-1 -ARLLPVRFRAPD-------GSLVATDLDLPAAPE-----WDAAGHGSYG
Mb1402c|M.bovis_AF2122/97 -RRAATMYRLDEQ-------DRLRHDVMGPPHVTP---PSFCNAGGGLWS
Rv1367c|M.tuberculosis_H37Rv -RRAATMYRLDEQ-------DRLRHDVMGPPHVTP---PSFCNAGGGLWS
MMAR_2187|M.marinum_M -RRAATMYRLDEL-------DQLRHDVMGPPHVRP---PSFCNAGGGLWS
MUL_1772|M.ulcerans_Agy99 -RRAATMYRLDEL-------DQLRHDVMGPPHVRP---PSFCNAGGGLWS
MAV_3395|M.avium_104 -HRAATMYSLDEH-------SRLQHGVMGPPHVTP---PAFCNAGGGLWS
MSMEG_3040|M.smegmatis_MC2_155 -RRAATMYKLTAE-------NTLTHDVMGPPPITD---PPFCAGGAGLFS
MAB_2833|M.abscessus_ATCC_1997 -ARLARLYGLGDD-------DTIVAADSGYLTAMPTSAPALCRGGGALAS
TH_4016|M.thermoresistible__bu RARIAPTSCDGDTPDLNPHRGRLLRGTVHDPTARR---MGGVSGSAGVFS
MLBr_00119|M.leprae_Br4923 -RPLPPIIAAIFR----KAIGGTVHEVIPYTNTPP--FLTTIVPSSNTVS
. . .
Mflv_3473|M.gilvum_PYR-GCK TVADYGRFVRAWLNDGELDGTRILKAETVELALRDHLDGAPLP-------
Mvan_1072|M.vanbaalenii_PYR-1 TVADYGRFVRAWLDGGELDGTRILKEETVELALRDHLDGAPLP-------
Mb1402c|M.bovis_AF2122/97 TADDYLRFVRMLLGDGTVDGVRVLSPESVRLMRTDRLT------------
Rv1367c|M.tuberculosis_H37Rv TADDYLRFVRMLLGDGTVDGVRVLSPESVRLMRTDRLT------------
MMAR_2187|M.marinum_M TADDYLRFVRLLLGDGTIDGVRVLSPESVRLMRTDRLS------------
MUL_1772|M.ulcerans_Agy99 TADDYLRFVRLLLGDGTIDGVRVLSPESVRLMRTDRLS------------
MAV_3395|M.avium_104 SADDYLRFVRMLLGDGTIDGVRVLSEESVRLMRTDRLT------------
MSMEG_3040|M.smegmatis_MC2_155 TVDDYLVFARMLLGGGTVDGVRVLSEESVRTMRTDRLT------------
MAB_2833|M.abscessus_ATCC_1997 TAHDYLTFARALLGGGQADGVRILSPESTQALRTNRLT------------
TH_4016|M.thermoresistible__bu TAADVGSYAQALLDRLAGRPSRFPLRRSTLQMMTSPQQPGHAPGQLDVAN
MLBr_00119|M.leprae_Br4923 TANELSRFAEIWLRGGELDGIRVLRSETLLGAVAECRR------------
:. : :.. * *. .: .
Mflv_3473|M.gilvum_PYR-GCK ---TKMEPTVPELAKAVELLPVPQGWGLG-FHLYH-----VDLPGMRSAG
Mvan_1072|M.vanbaalenii_PYR-1 ---KKMEPAVPELSNPVELLDVPQGWGLG-FHLYL-----IDLPGMRSAG
Mb1402c|M.bovis_AF2122/97 -------DEQKRHSFLGAPFWVGRGFGLN-LSVVTDPAKSRPLFGPGGLG
Rv1367c|M.tuberculosis_H37Rv -------DEQKRHSFLGAPFWVGRGFGLN-LSVVTDPAKSRPLFGPGGLG
MMAR_2187|M.marinum_M -------DEHKRHNFLGAPFWVGRGFGLN-LSVVTDPAQSTPLFGPGGLG
MUL_1772|M.ulcerans_Agy99 -------DEHKRHNFLGAPFWVGRGFGLN-LSVVTDPVQSTPLFGPGGLG
MAV_3395|M.avium_104 -------EAQKRHNFLGAPYWVGRGFGLN-LSVVTDPARSIPLFGPGGVG
MSMEG_3040|M.smegmatis_MC2_155 -------PEQKQYPFLGAPFWVGRGFGLN-LSVVTDPAKSRQLFGPGGLG
MAB_2833|M.abscessus_ATCC_1997 -------PAQRRLPSFGIPYWTGRGFGLG-LSVVMDPNE-AALFGPGGTG
TH_4016|M.thermoresistible__bu KAAREAVLSASDDPLLAPDYPAVPGRDLRGFGWDIDTNQSRPRGSVFPVG
MLBr_00119|M.leprae_Br4923 --------LRPDFVVGLMPARWGTGFILG--------TNKFGPFGRNAPA
* * . .
Mflv_3473|M.gilvum_PYR-GCK SGDWSGLFNSFFWIDRTAGIGGVIATQL-LPFFD-------------DTM
Mvan_1072|M.vanbaalenii_PYR-1 SADWSGLFNSFFWIDRKAGIGAVIATQL-LPFFD-------------GKM
Mb1402c|M.bovis_AF2122/97 TFSWPGAYGTWWQADPSADLILLYLIQH-CPDLSVDAAAAVAGNPSLAKL
Rv1367c|M.tuberculosis_H37Rv TFSWPGAYGTWWQADPSADLILLYLIQH-CPDLSVDAAAAVAGNPSLAKL
MMAR_2187|M.marinum_M TFSWPGAYGTWWQADPGADLILLYLIQH-CPDLSVNAAAAVAGNPGLAKL
MUL_1772|M.ulcerans_Agy99 TFSWPGAYGTWWQADPGADLILLYLIQH-CPDLSVNAAAAVAGNPGLAKL
MAV_3395|M.avium_104 TFSWPGAYGTWWQADPTAELILIYLIQN-LPDLTVDAATAVAGNTSLAKL
MSMEG_3040|M.smegmatis_MC2_155 TFSWPGAYGTWWQADPSADLILIYLIQN-FPDLGVDAA-AIAGNTSLAKL
MAB_2833|M.abscessus_ATCC_1997 TFGWPGAFGTWWHADPKADAILMFLPQWRMPELDPKAALARTST---IRL
TH_4016|M.thermoresistible__bu SFGHTGFTGVTLWIDPGSDTYVVVLANVIHQPGGPP----------VAPL
MLBr_00119|M.leprae_Br4923 AFGHLGLVNIAIWADPQRSMAAGLISSG-KPGQDPN----------AGRY
: . * . * .
Mflv_3473|M.gilvum_PYR-GCK VETILAFEAAVYASL----
Mvan_1072|M.vanbaalenii_PYR-1 VETILRFEATVYAELGAGV
Mb1402c|M.bovis_AF2122/97 RTAQPKFVRRTYRALGL--
Rv1367c|M.tuberculosis_H37Rv RTAQPKFVRRTYRALGL--
MMAR_2187|M.marinum_M RTAQPRFVRRTYRALGL--
MUL_1772|M.ulcerans_Agy99 RTAQPRFVRRTYRALGL--
MAV_3395|M.avium_104 RTAQPRFVRTTYRALGL--
MSMEG_3040|M.smegmatis_MC2_155 QSAQPKFVRRTYQALDL--
MAB_2833|M.abscessus_ATCC_1997 QLLHVQFGQAVYAAL----
TH_4016|M.thermoresistible__bu SGRVATLAARALHLYGT--
MLBr_00119|M.leprae_Br4923 GALMNTITAEIPVH-----
: