For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVTRVLIADDDGVVRDVVRRYLERDGLEVSTAHDGNEALRLLSSQRIDVAVLDVMMPGPDGMALCRRLRQ NGHYSVPVILLTALGEEDDRIAGLEAGADDYLTKPFSPRELALRVRSVLRRSPASIAAHPVDITSGDLTV STASRSATVGGRTVSLTNREFDLLLFFLTHAGTVYTREDLLKEVWHWDFGDLSTVTVHVKRLRSKLGDRH RVQTVWGRGYMWAPDEASPGQPDAAG
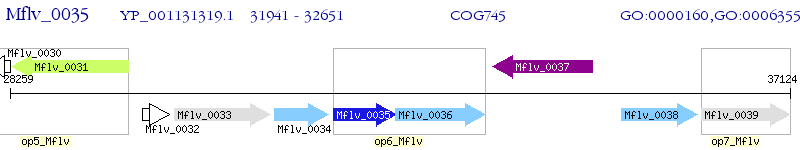
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0035 | - | - | 100% (236) | two component transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1887 | - | 5e-44 | 44.25% (226) | two component transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0461 | - | 9e-41 | 47.06% (221) | two component transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1007 | mprA | 3e-43 | 43.11% (225) | two component response transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv0981 | mprA | 6e-43 | 43.11% (225) | two component response transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_00174 | - | 2e-42 | 42.79% (222) | putative two-component response regulator |
| M. abscessus ATCC 19977 | MAB_1076 | - | 1e-42 | 42.22% (225) | two component response transcriptional regulatory protein MprA |
| M. marinum M | MMAR_4529 | mprA | 1e-42 | 43.75% (224) | two component response transcriptional regulatory protein |
| M. avium 104 | MAV_1094 | - | 8e-44 | 44.14% (222) | two component response transcriptional regulatory protein MprA |
| M. smegmatis MC2 155 | MSMEG_0994 | resD | 5e-99 | 76.62% (231) | DNA-binding response regulator ResD |
| M. thermoresistible (build 8) | TH_1732 | phoP | 3e-43 | 42.67% (225) | DNA-binding response regulator PhoP |
| M. ulcerans Agy99 | MUL_4701 | mprA | 8e-43 | 43.75% (224) | two component response transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0885 | - | 1e-104 | 84.30% (223) | two component transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1007|M.bovis_AF2122/97 MSVRILVVDDDRAVRESLRRSLSFNGYSVELAHDGVEALDMIASDRPDAL
Rv0981|M.tuberculosis_H37Rv MSVRILVVDDDRAVRESLRRSLSFNGYSVELAHDGVEALDMIASDRPDAL
MAV_1094|M.avium_104 --MRILVVDDDRAVRESLRRSLSFNGYSVELAHDGVEALDMIASDRPDAL
MMAR_4529|M.marinum_M MSVRILVVDDDRAVRESLRRSLSFNGYTVELAHDGVEALEMIASDRPDAL
MUL_4701|M.ulcerans_Agy99 MSVRILVVDDDRAVRESLRRSLSFNGYTVELAHDGVEALEMIASDRPDAL
MLBr_00174|M.leprae_Br4923 --MRILAVDDDRAVRESLRRSLSFNGYSVELANDGVEALEMVARDRPDAL
TH_1732|M.thermoresistible__bu VAVRILVVDDDRAVRESLRRSLSFNGYSVELAEDGVEALDLIANDRPDAV
MAB_1076|M.abscessus_ATCC_1997 MPVRILVVDDDRAVRESLRRSLSFNGYSVDLAEDGVDALNVIANDRPDAL
Mflv_0035|M.gilvum_PYR-GCK MVTRVLIADDDGVVRDVVRRYLERDGLEVSTAHDGNEALRLLSSQRIDVA
Mvan_0885|M.vanbaalenii_PYR-1 ----MLIADDDDVVRDVVRRYLERDGLEVCTAPNGSEALRLLGSRRIDVA
MSMEG_0994|M.smegmatis_MC2_155 -MPRVLIADDDTVVRDVVRRYLERDGLDVTIAHDGSEALRLLESEQLDVA
:* .*** .**: :** *. :* * * :* :** :: : *.
Mb1007|M.bovis_AF2122/97 VLDVMMPRLDGLEVCRQLRSTGDD-LPILVLTARDSVSERVAGLDAGADD
Rv0981|M.tuberculosis_H37Rv VLDVMMPRLDGLEVCRQLRGTGDD-LPILVLTARDSVSERVAGLDAGADD
MAV_1094|M.avium_104 VLDVMMPRLDGLEVCRQLRSTGDD-LPILVLTARDSVSERVAGLDAGADD
MMAR_4529|M.marinum_M VLDVMMPRLDGLEVCRQLRSTGDD-LPILVLTARDSVSERVAGLDAGADD
MUL_4701|M.ulcerans_Agy99 VLDVMMPRLDGLEVCRQLRSTGDD-LPILVLTARDSVSERVAGLDAGADD
MLBr_00174|M.leprae_Br4923 VLDVMMPRLDGLEVCRQLRSTGDD-LPILVLTARDSVSERVAGLDAGADD
TH_1732|M.thermoresistible__bu VLDVMMPRLDGLEVCRQLRSTGDD-LPILVLTARDSVSERVAGLDAGADD
MAB_1076|M.abscessus_ATCC_1997 VLDVMMPKLDGLEVCRRLRSTGDD-LPILVLTARDSVSERVAGLDAGADD
Mflv_0035|M.gilvum_PYR-GCK VLDVMMPGPDGMALCRRLRQNGHYSVPVILLTALGEEDDRIAGLEAGADD
Mvan_0885|M.vanbaalenii_PYR-1 VLDVMMPGPDGIALCRNLRKGGDYSVPVILLTALGEEQDRIAGLEAGADD
MSMEG_0994|M.smegmatis_MC2_155 VLDVMMPGPDGLSLCRELRKYGDYRLPVILLTALGEEDDRIAGLEAGADD
******* **: :**.** *. :*:::*** .. .:*:***:*****
Mb1007|M.bovis_AF2122/97 YLPKPFALEELLARMRALLRRTKPED--AAESMAMRFSDLTLDPVTREVN
Rv0981|M.tuberculosis_H37Rv YLPKPFALEELLARMRALLRRTKPED--AAESMAMRFSDLTLDPVTREVN
MAV_1094|M.avium_104 YLPKPFALEELLARMRALLRRTKPED--DAESVAMTFSDLTLDPVTREVT
MMAR_4529|M.marinum_M YLPKPFALEELLARMRALLRRTKPEDDEAADSVAMTFSDLSLDPVTREVT
MUL_4701|M.ulcerans_Agy99 YLPKPFALEELLARMRALLRRTKLEDDEAADSVAMTFSDLSLDPVTREVT
MLBr_00174|M.leprae_Br4923 YLPKPFALEELLARIRALLRRTKPDD--AAESVAMSFSDLTLDPVTREVA
TH_1732|M.thermoresistible__bu YLPKPFALEELLARLRALLRRTTPDD--AADSQKLTFADLTLDPVTREVT
MAB_1076|M.abscessus_ATCC_1997 YLPKPFALEELLARMRALLRRTTIDE--SADAATLSFGDLTLDPVTREVH
Mflv_0035|M.gilvum_PYR-GCK YLTKPFSPRELALRVRSVLRRSPASI--AAHPVDITSGDLTVSTASRSAT
Mvan_0885|M.vanbaalenii_PYR-1 YLTKPFSPRELALRVRSVLRRSPATG--GVLPVDIRSGDLTVTTASRTVT
MSMEG_0994|M.smegmatis_MC2_155 YLTKPFSPRELALRVRSVLRRTSGLT--TPAPAELTVGELRVSTASRSVT
**.***: .** *:*::***: . : .:* : ..:* .
Mb1007|M.bovis_AF2122/97 RGQRRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEV
Rv0981|M.tuberculosis_H37Rv RGQRRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEV
MAV_1094|M.avium_104 RGQRRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEV
MMAR_4529|M.marinum_M RGARRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEV
MUL_4701|M.ulcerans_Agy99 RGARRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEV
MLBr_00174|M.leprae_Br4923 RGQRWISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEV
TH_1732|M.thermoresistible__bu RGNRQISLTRTEFALLELLIANPRRVLSRSRILEEVWGFDFPTSGNALEV
MAB_1076|M.abscessus_ATCC_1997 RGTRSISLTRTEFALLEMLMANPRRVLTRSRILEEVWGFDFPTSGNALEV
Mflv_0035|M.gilvum_PYR-GCK VGGRTVSLTNREFDLLLFFLTHAGTVYTREDLLKEVWHWDFGDLS-TVTV
Mvan_0885|M.vanbaalenii_PYR-1 MAGTPVSLTNREFDLLLFFLTHPDTVFTREDLLKQVWLWDFGDLS-TVTV
MSMEG_0994|M.smegmatis_MC2_155 LSGRPVNLTNREFDLLVFFLTHTDTVFSREDLLKRVWRWDFGDLS-TVTV
. :.**. ** ** :::::. * :*. :*:.** :** . :: *
Mb1007|M.bovis_AF2122/97 YVGYLRRKTEADGEPRLIHTVRGVGYVLR---------ETPP
Rv0981|M.tuberculosis_H37Rv YVGYLRRKTEADGEPRLIHTVRGVGYVLR---------ETPP
MAV_1094|M.avium_104 YVGYLRRKTEADGEPRLIHTVRGVGYVLR---------ETPP
MMAR_4529|M.marinum_M YVGYLRRKTEAEGEPRLIHTVRGVGYVLR---------ETPP
MUL_4701|M.ulcerans_Agy99 YVGYLRRKTEAEGEPRLIHTVRGVGYVLR---------ETPP
MLBr_00174|M.leprae_Br4923 YVGYLRRKTEADGESRLIHTVRGVGYVLR---------ETPP
TH_1732|M.thermoresistible__bu YIGYLRRKTEAEGEPRLIHTVRGVGYVLR---------ETPP
MAB_1076|M.abscessus_ATCC_1997 YVGYLRRKTEAEGELRLIHTVRGVGYVLR---------ETPP
Mflv_0035|M.gilvum_PYR-GCK HVKRLRSK---LGDRHRVQTVWGRGYMWAPDEASPGQPDAAG
Mvan_0885|M.vanbaalenii_PYR-1 HVKRLRSK---LGDRHRVQTVWGRGYMWT--------SGAAG
MSMEG_0994|M.smegmatis_MC2_155 HVKRLRSK---LGTHHRVQTVWGRGYLWS---------GQP-
:: ** * * : ::** * **: .