For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SVRILVVDDDRAVRESLRRSLSFNGYTVELAHDGVEALEMIASDRPDALVLDVMMPRLDGLEVCRQLRST GDDLPILVLTARDSVSERVAGLDAGADDYLPKPFALEELLARMRALLRRTKPEDDEAADSVAMTFSDLSL DPVTREVTRGARRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVYVGYLRRKTEAE GEPRLIHTVRGVGYVLRETPP*
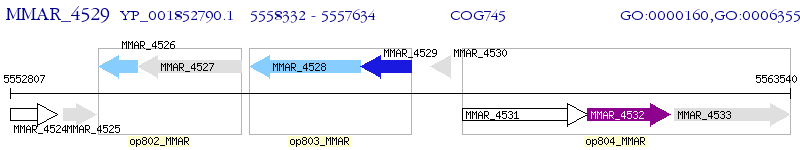
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4529 | mprA | - | 100% (232) | two component response transcriptional regulatory protein MprA |
| M. marinum M | MMAR_4455 | trcR | 1e-58 | 49.12% (226) | two-component transcriptional regulator TrcR |
| M. marinum M | MMAR_4633 | prrA | 6e-57 | 50.22% (227) | two-component response transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1007 | mprA | 1e-120 | 94.83% (232) | two component response transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_1887 | - | 1e-115 | 91.74% (230) | two component transcriptional regulator |
| M. tuberculosis H37Rv | Rv0981 | mprA | 1e-120 | 94.40% (232) | two component response transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_00174 | - | 1e-116 | 92.61% (230) | putative two-component response regulator |
| M. abscessus ATCC 19977 | MAB_1076 | - | 1e-111 | 88.70% (230) | two component response transcriptional regulatory protein MprA |
| M. avium 104 | MAV_1094 | - | 1e-122 | 96.52% (230) | two component response transcriptional regulatory protein MprA |
| M. smegmatis MC2 155 | MSMEG_5488 | - | 1e-114 | 89.66% (232) | DNA-binding response regulator |
| M. thermoresistible (build 8) | TH_1732 | phoP | 1e-116 | 90.09% (232) | DNA-binding response regulator PhoP |
| M. ulcerans Agy99 | MUL_4701 | mprA | 1e-127 | 99.57% (232) | two component response transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_4844 | - | 1e-117 | 93.48% (230) | two component transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1887|M.gilvum_PYR-GCK MPVRILVVDDDRAVRESLRRSLSFNGYSVELAQDGREALDLIASDRPDAV
Mvan_4844|M.vanbaalenii_PYR-1 MPVRILVVDDDRAVRESLRRSLSFNGYSVELAQDGREALDLIASDRPDAV
MMAR_4529|M.marinum_M MSVRILVVDDDRAVRESLRRSLSFNGYTVELAHDGVEALEMIASDRPDAL
MUL_4701|M.ulcerans_Agy99 MSVRILVVDDDRAVRESLRRSLSFNGYTVELAHDGVEALEMIASDRPDAL
Mb1007|M.bovis_AF2122/97 MSVRILVVDDDRAVRESLRRSLSFNGYSVELAHDGVEALDMIASDRPDAL
Rv0981|M.tuberculosis_H37Rv MSVRILVVDDDRAVRESLRRSLSFNGYSVELAHDGVEALDMIASDRPDAL
MAV_1094|M.avium_104 --MRILVVDDDRAVRESLRRSLSFNGYSVELAHDGVEALDMIASDRPDAL
MLBr_00174|M.leprae_Br4923 --MRILAVDDDRAVRESLRRSLSFNGYSVELANDGVEALEMVARDRPDAL
MSMEG_5488|M.smegmatis_MC2_155 MAVRILVVDDDRAVRESLRRSLSFNGYSVELAQDGVEALDAITNNRPDAL
TH_1732|M.thermoresistible__bu VAVRILVVDDDRAVRESLRRSLSFNGYSVELAEDGVEALDLIANDRPDAV
MAB_1076|M.abscessus_ATCC_1997 MPVRILVVDDDRAVRESLRRSLSFNGYSVDLAEDGVDALNVIANDRPDAL
:***.********************:*:**.** :**: :: :****:
Mflv_1887|M.gilvum_PYR-GCK VLDVMMPRLDGLEVCRQLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
Mvan_4844|M.vanbaalenii_PYR-1 VLDVMMPRLDGLEVCRQLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
MMAR_4529|M.marinum_M VLDVMMPRLDGLEVCRQLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
MUL_4701|M.ulcerans_Agy99 VLDVMMPRLDGLEVCRQLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
Mb1007|M.bovis_AF2122/97 VLDVMMPRLDGLEVCRQLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
Rv0981|M.tuberculosis_H37Rv VLDVMMPRLDGLEVCRQLRGTGDDLPILVLTARDSVSERVAGLDAGADDY
MAV_1094|M.avium_104 VLDVMMPRLDGLEVCRQLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
MLBr_00174|M.leprae_Br4923 VLDVMMPRLDGLEVCRQLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
MSMEG_5488|M.smegmatis_MC2_155 ILDVMMPRLDGLEVCRQLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
TH_1732|M.thermoresistible__bu VLDVMMPRLDGLEVCRQLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
MAB_1076|M.abscessus_ATCC_1997 VLDVMMPKLDGLEVCRRLRSTGDDLPILVLTARDSVSERVAGLDAGADDY
:******:********:**.******************************
Mflv_1887|M.gilvum_PYR-GCK LPKPFALEELLARMRALLRRTSPVDG-DGESAAMTFSDLTLDPVTREVTR
Mvan_4844|M.vanbaalenii_PYR-1 LPKPFALEELLARMRALLRRTTPDDG-AGESAAMTFSDLSLDPVTREVTR
MMAR_4529|M.marinum_M LPKPFALEELLARMRALLRRTKPEDDEAADSVAMTFSDLSLDPVTREVTR
MUL_4701|M.ulcerans_Agy99 LPKPFALEELLARMRALLRRTKLEDDEAADSVAMTFSDLSLDPVTREVTR
Mb1007|M.bovis_AF2122/97 LPKPFALEELLARMRALLRRTKPED--AAESMAMRFSDLTLDPVTREVNR
Rv0981|M.tuberculosis_H37Rv LPKPFALEELLARMRALLRRTKPED--AAESMAMRFSDLTLDPVTREVNR
MAV_1094|M.avium_104 LPKPFALEELLARMRALLRRTKPED--DAESVAMTFSDLTLDPVTREVTR
MLBr_00174|M.leprae_Br4923 LPKPFALEELLARIRALLRRTKPDD--AAESVAMSFSDLTLDPVTREVAR
MSMEG_5488|M.smegmatis_MC2_155 LPKPFALEELLARMRALLRRTVSDD--SGDSQKMTFSDLTLDPVTREVTR
TH_1732|M.thermoresistible__bu LPKPFALEELLARLRALLRRTTPDD--AADSQKLTFADLTLDPVTREVTR
MAB_1076|M.abscessus_ATCC_1997 LPKPFALEELLARMRALLRRTTIDE--SADAATLSFGDLTLDPVTREVHR
*************:******* : .:: : *.**:******** *
Mflv_1887|M.gilvum_PYR-GCK GNRSISLTRTEFSLLEMLITNPRRVLTRSRILEEVWGFDFPTSGNALEVY
Mvan_4844|M.vanbaalenii_PYR-1 GDRPISLTRTEFSLLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVY
MMAR_4529|M.marinum_M GARRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVY
MUL_4701|M.ulcerans_Agy99 GARRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVY
Mb1007|M.bovis_AF2122/97 GQRRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVY
Rv0981|M.tuberculosis_H37Rv GQRRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVY
MAV_1094|M.avium_104 GQRRISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVY
MLBr_00174|M.leprae_Br4923 GQRWISLTRTEFALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVY
MSMEG_5488|M.smegmatis_MC2_155 GGRQISLTRTEFSLLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVY
TH_1732|M.thermoresistible__bu GNRQISLTRTEFALLELLIANPRRVLSRSRILEEVWGFDFPTSGNALEVY
MAB_1076|M.abscessus_ATCC_1997 GTRSISLTRTEFALLEMLMANPRRVLTRSRILEEVWGFDFPTSGNALEVY
* * ********:***:*::******:***********************
Mflv_1887|M.gilvum_PYR-GCK VGYLRRKTEAEGEPRLIHTVRGVGYVLRETPP
Mvan_4844|M.vanbaalenii_PYR-1 VGYLRRKTEAEGEPRLIHTVRGVGYVLRETPP
MMAR_4529|M.marinum_M VGYLRRKTEAEGEPRLIHTVRGVGYVLRETPP
MUL_4701|M.ulcerans_Agy99 VGYLRRKTEAEGEPRLIHTVRGVGYVLRETPP
Mb1007|M.bovis_AF2122/97 VGYLRRKTEADGEPRLIHTVRGVGYVLRETPP
Rv0981|M.tuberculosis_H37Rv VGYLRRKTEADGEPRLIHTVRGVGYVLRETPP
MAV_1094|M.avium_104 VGYLRRKTEADGEPRLIHTVRGVGYVLRETPP
MLBr_00174|M.leprae_Br4923 VGYLRRKTEADGESRLIHTVRGVGYVLRETPP
MSMEG_5488|M.smegmatis_MC2_155 IGYLRRKTEAEGEPRLIHTVRGVGYVLRETPP
TH_1732|M.thermoresistible__bu IGYLRRKTEAEGEPRLIHTVRGVGYVLRETPP
MAB_1076|M.abscessus_ATCC_1997 VGYLRRKTEAEGELRLIHTVRGVGYVLRETPP
:*********:** ******************