For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGGLRFGFVDALVHSRLPPTLPARSSMAAATVMGADSYWVGDHLNALVPRSIATSEYLGIAAKFVPKIDA NYEPWTMLGNLAFGLPSRLRLGVCVTDAGRRNPAVTAQAAATLHLLTRGRAILGIGVGEREGNEPYGVEW TKPVARFEEALATIRALWNSNGELISRESPYFPLHNALFDLPPYRGKWPEIWVAAHGPRMLRATGRYADA WIPIVVVRPSDYSRALEAVRSAASDAGRDPMSITPAAVRGIITGRNRDDVEEALESVVVKMTALGVPGEA WARHGVEHPMGADFSGVQDIIPQTMDKQTVLSYAAKVPAALMKEVVFSGTPDEVIDQVAEWRDHGLRYVV LINGSLVNPSLRKTVTAVLPHAKVLRGLKKL
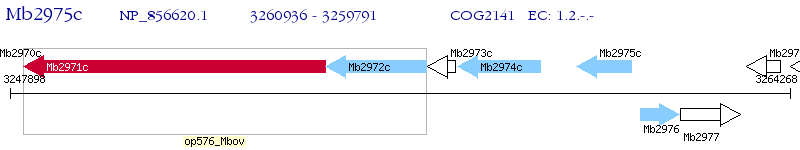
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2975c | - | - | 100% (381) | oxidoreductase |
| M. bovis AF2122 / 97 | Mb0815c | - | 1e-15 | 24.92% (313) | hypothetical protein Mb0815c |
| M. bovis AF2122 / 97 | Mb0137c | fgd2 | 1e-15 | 33.98% (206) | putative f420-dependant glucose-6-phosphate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3380 | - | 1e-163 | 71.35% (377) | luciferase family protein |
| M. tuberculosis H37Rv | Rv2951c | - | 0.0 | 100.00% (381) | oxidoreductase |
| M. leprae Br4923 | MLBr_00131 | - | 0.0 | 85.08% (382) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0858 | - | 2e-17 | 24.69% (320) | luciferase-like monooxygenase |
| M. marinum M | MMAR_1758 | - | 0.0 | 83.68% (380) | ketoreductase |
| M. avium 104 | MAV_0066 | - | 3e-56 | 38.92% (316) | hypothetical protein MAV_0066 |
| M. smegmatis MC2 155 | MSMEG_6454 | - | 5e-17 | 33.18% (211) | hypothetical protein MSMEG_6454 |
| M. thermoresistible (build 8) | TH_2487 | - | 3e-17 | 32.63% (190) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3202 | - | 5e-18 | 30.80% (263) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_2018 | - | 1e-15 | 31.38% (188) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2975c|M.bovis_AF2122/97 ----------MGG--LRFGFVDALVHSRLPPTLPARSSMAAATVMG-ADS
Rv2951c|M.tuberculosis_H37Rv ----------MGG--LRFGFVDALVHSRLPPTLPARSSMAAATVMG-ADS
MLBr_00131|M.leprae_Br4923 ----------MAG--FRFGFVDALVHTLFPPSLPARASIVSGAVLG-ADS
MMAR_1758|M.marinum_M -------MKKSSG--FRYGFVDAPVHSRLPPSMIARSSMMAGAVMG-ADS
Mflv_3380|M.gilvum_PYR-GCK ----MSPRPRSAGSQFRFGLLDGIVNTRVSPTLLPSAAMLTASATG-ADS
MAV_0066|M.avium_104 --------------------MTVQPAAFLRTTLPLDLSRLDEFDSGRYHS
MSMEG_6454|M.smegmatis_MC2_155 MRFAFKTSPQN------------------TTWADMLAIWKAADNIDVFES
MUL_3202|M.ulcerans_Agy99 MHENTSNPPQHGGPQFGLILALMTQGLAISDYPDMVRVSTLAESYG-FDS
MAB_0858|M.abscessus_ATCC_1997 --MAQVAETLEFGVFIPQGWRLDLVGIAPERHWDVMRDLVHYAEGGPWDS
TH_2487|M.thermoresistible__bu -----------------------MTDYGHDLLFGTFVTPSNHPPHHPVEQ
Mvan_2018|M.vanbaalenii_PYR-1 -----------------------MTRVLDELGFYLLAGAGGEGPATLMDE
..
Mb2975c|M.bovis_AF2122/97 YWVGDHLNALVPRSIATS-----EYLGIAAKFVPKIDANYEPWTMLGNLA
Rv2951c|M.tuberculosis_H37Rv YWVGDHLNALVPRSIATS-----EYLGIAAKFVPKIDANYEPWTMLGNLA
MLBr_00131|M.leprae_Br4923 YWVGDHLNALVPRSVATP-----KYLGVAAKVVPKIDANYEPWTMLGNLA
MMAR_1758|M.marinum_M YWIGDHLNALVPRSVATP-----EFLGLGARLVPKVDANYEPWTMLGNLA
Mflv_3380|M.gilvum_PYR-GCK FWVGDHINSLVPRSVATP-----EHLGLAAKLVPKVDAILEPWTMLGHLA
MAV_0066|M.avium_104 VWLPDHMVSFWPDSIWAP-----EFTDLATASASPHRHLDG---LAVAAA
MSMEG_6454|M.smegmatis_MC2_155 GWTFDHFYPIFSD---------------------SAGPCLEGWTTLTALA
MUL_3202|M.ulcerans_Agy99 VWLCDHFLTLSPDDYVTQAGISGEHERRDGRDRPGTMPLLEAWTALSALA
MAB_0858|M.abscessus_ATCC_1997 VWVYDHFHTVPVP---------------------TDEATHEAWTLMSAFA
TH_2487|M.thermoresistible__bu AVVADRAGLDLVTFQDHP----------------YVAKYHDTWTLMSYAA
Mvan_2018|M.vanbaalenii_PYR-1 ARRGEELGFGTGFISER-------------------WNVKEASSLTGAAL
:.
Mb2975c|M.bovis_AF2122/97 FG-LPSRLRLGVCVTDAGRRNPAVTAQAAATLHLLTRGRAILGIGVGERE
Rv2951c|M.tuberculosis_H37Rv FG-LPSRLRLGVCVTDAGRRNPAVTAQAAATLHLLTRGRAILGIGVGERE
MLBr_00131|M.leprae_Br4923 AGNRLNGLRLGVCVTDAGRRNPAVTAQAAATLHLLTRGKAMLGIGVGERE
MMAR_1758|M.marinum_M AGNRINRLRLGVCVTDVGRRNPAVTAQAAATVHLLTRGRAILGIGVGERE
Mflv_3380|M.gilvum_PYR-GCK AR-HRGRLRFGVGVTDASRRHPAVTAQAAATLHLLTRGRAILGIGVGERE
MAV_0066|M.avium_104 AAVLTERVPLVSAVVDTVRRHPALLAQTALTLDHLSKGRFILGLGSGESE
MSMEG_6454|M.smegmatis_MC2_155 QA--TERLRVGVLVTGIHYRHPAVLANMAAALDIISNGRLELGIGAGWNE
MUL_3202|M.ulcerans_Agy99 RD--TTYIRLGTSVLCNSYRHPAVLAKMAATLDIISEGRLDLGLGAGWFE
MAB_0858|M.abscessus_ATCC_1997 AV--TSRIKLGQMCTAMGYRNPVYLAKIAATVDIISGGRAQMGIGGGWYE
TH_2487|M.thermoresistible__bu AR--TQRVRLSANVANLPLRPPAVLARSAAGLDLLSGGRVELGLGAGAFW
Mvan_2018|M.vanbaalenii_PYR-1 AV--TSRMQIATAATNHNTRHPLITGSWATTMHRLSGGRFTLGIGRG-IG
: . * * . * :. :: *: :*:* *
Mb2975c|M.bovis_AF2122/97 GN-EPYGVEWT---KPVARFEEALATIRALWNSNGE-LISRESPYFPLHN
Rv2951c|M.tuberculosis_H37Rv GN-EPYGVEWT---KPVARFEEALATIRALWNSNGE-LISRESPYFPLHN
MLBr_00131|M.leprae_Br4923 GN-EPYGVEWT---KPVARFQEALATIRALWDSNGE-LVSRESQFFPLHN
MMAR_1758|M.marinum_M GN-EPYGVEWS---KPVARFEEGLATIRALWDSNGA-LVTRESPYFPLRN
Mflv_3380|M.gilvum_PYR-GCK GN-EPYGVEWT---KPVARFVEALATIRALWESGGE-PVTRESDYFPLRH
MAV_0066|M.avium_104 NT-VPYGFDFA---RPVSRFEEALRVIRLLWDTDG--PVDFNGEFYSLHQ
MSMEG_6454|M.smegmatis_MC2_155 EESGAYGIELGSIKERFDRFEEACEVLKGLLTQET---TTFDGKFYQLKD
MUL_3202|M.ulcerans_Agy99 REFTAYGIPFLPVGDRVTGLGEALEVITAIWTQPD---PVYSGRFYTLDG
MAB_0858|M.abscessus_ATCC_1997 HEWRAYGYGFPGAAERLGRLREGVRIMRDAWRDGR---VSLDGTYYQVDD
TH_2487|M.thermoresistible__bu DGVVAMGGERRSPGAAIKALEEAIAVIRGIWDVDAAGVLDVDGAHYRIRG
Mvan_2018|M.vanbaalenii_PYR-1 AIYNAFGIPTV----TTAQMEDFAQVMRKLWHGELI--FNHDGPLGKYQV
. * : : : ..
Mb2975c|M.bovis_AF2122/97 ALFDLPPYRGKWPEIWVAAHGP-RMLRATGRYADAWIPIVVVRP---SDY
Rv2951c|M.tuberculosis_H37Rv ALFDLPPYRGKWPEIWVAAHGP-RMLRATGRYADAWIPIVVVRP---SDY
MLBr_00131|M.leprae_Br4923 ALFDLPPYRGKWPEIWVAAHGP-RMLQATGRYADAWIPIVLVRP---TDY
MMAR_1758|M.marinum_M ALFDLPPYKGKWPEIWVAAHGP-RMLRATGRYADAWVPIVIIRP---TDY
Mflv_3380|M.gilvum_PYR-GCK AVFDLPPYKGRWPEIWVAAHGP-RMLRATGRYADGWVPFVISRP---ADY
MAV_0066|M.avium_104 ARLDTEPYHDRLPPIWIGASGP-RMLDIAGRYADGWWPAGAWTP---EHY
MSMEG_6454|M.smegmatis_MC2_155 ARNEPKGPQQPHPPFCIGGSGEKRTLRITAKYADHWN-FVGGTP---EEF
MUL_3202|M.ulcerans_Agy99 AICDPPPVQTPHPPLWVGGEGD-RVHRIAARSAQGIN-VRWWLP---QKV
MAB_0858|M.abscessus_ATCC_1997 AIVAPTPLQSNGIPLWIAGGGEKVTLRIAARYAQYTN--FGQTA---EEF
TH_2487|M.thermoresistible__bu AKRGPAPAHR--ISIWIGGYGP-RMLGLVGRLADGWLPSLQYLSGGVSDL
Mvan_2018|M.vanbaalenii_PYR-1 LFLD--PDFREDIRLAIVAFGP-QTLALGGREFDDVILHTYFTP---ETL
: : . * .: : .
Mb2975c|M.bovis_AF2122/97 SRALEAVRSAASDAGRDPMSITPAAVRGIITGRNRDDVEEALESVVVKMT
Rv2951c|M.tuberculosis_H37Rv SRALEAVRSAASDAGRDPMSITPAAVRGIITGRNRDDVEEALESVVVKMT
MLBr_00131|M.leprae_Br4923 SCALEVVRTAASDAGRDPMSITPAAVRGIITGRTRDDVDEALDSVLVRMI
MMAR_1758|M.marinum_M SRALEAVRAAASDAGRDPMSITPSAVRGVVTGWSRDDVDEALDSVIVKMN
Mflv_3380|M.gilvum_PYR-GCK ARALDAVRTAASDNGRDPMAIVPAVNRTVVTGRTRADVDEALDSVIVKSV
MAV_0066|M.avium_104 AEMLGAVRVSAERAGRDPMAITPCFIQVCLIGEDDDALAEILRAPLVKAF
MSMEG_6454|M.smegmatis_MC2_155 ARKRDVLAAHCADLGRDPKEIT-----------------------LSAHV
MUL_3202|M.ulcerans_Agy99 KQRKDFLAQTCATAQRDPTTLR-----------------------FSVTA
MAB_0858|M.abscessus_ATCC_1997 GHKSRVLAAHCKDIGTDFDAIVRSANMNIVIGSDAG---------EVADR
TH_2487|M.thermoresistible__bu TAMNQRIDDAAVAAGRDPRQIR---------------------------R
Mvan_2018|M.vanbaalenii_PYR-1 QRAVRTVKDAAEQAGRDPASVRVWSCFATVGDHLPEELRLKKTVARLATY
: . * :
Mb2975c|M.bovis_AF2122/97 ALGVPGEAWARHGVEHPMGADFSGVQDII-----PQTMDKQTVLSYAAKV
Rv2951c|M.tuberculosis_H37Rv ALGVPGEAWARHGVEHPMGADFSGVQDII-----PQTMDKQTVLSYAAKV
MLBr_00131|M.leprae_Br4923 ALGVPGEAWARHGVEHPMGADFAGVQDII-----PQTIDEETVVSYAAKV
MMAR_1758|M.marinum_M ALGMPAEAWARHGVEHPMGADFTGVQDLI-----PQTMDRETVLSYTAKV
Mflv_3380|M.gilvum_PYR-GCK ALAAPAEAWARHGVEHPLGSDFSGVQDLV-----PQMIDKEAALAYTASV
MAV_0066|M.avium_104 LLQVSAKTLGEFGFEHPMGDGWRGFQDID-----PAALGRDRIVEFLHRV
MSMEG_6454|M.smegmatis_MC2_155 RLGEDRDYAKVIDEAAALGAEGLDLAIIY-----LPP-------PYDPAV
MUL_3202|M.ulcerans_Agy99 LLAPTDSAEQEAQIRTEFSAVPYDGLIVG-----PADRCIERINEYYESG
MAB_0858|M.abscessus_ATCC_1997 LAAIKARLVPIIGEDVAEGTVANLATTAG-----TPEQIAERLAEYRGLG
TH_2487|M.thermoresistible__bu LLNVGGQPSADELADIALSHGISAFILAS-----DDTATTERFAAEVAPA
Mvan_2018|M.vanbaalenii_PYR-1 LQGYGDLLVNTNGWDPAVLQRFRDDKVVQSVGGGIDHKATAEQIEHIATL
Mb2975c|M.bovis_AF2122/97 PAALMKEVVFSGTPDEVIDQVAEWRDHGLRYVVLINGSLVNPSLRKTVTA
Rv2951c|M.tuberculosis_H37Rv PAALMKEVVFSGTPDEVIDQVAEWRDHGLRYVVLINGSLVNPSLRKTVTA
MLBr_00131|M.leprae_Br4923 PAALMKEVLFSGTPEEVIDQVAEWRDHGLKYLVVINGSLVNSSLRKTVSA
MMAR_1758|M.marinum_M PPGLIREVVASGTPDEVIDQVAEWRDHGLRYLLVINGSLVNPSLRKTVTA
Mflv_3380|M.gilvum_PYR-GCK PASLMKEICFHGTADEVLDQVAEWRDHGLRYLLVINGSQLNPKLRKGVSA
MAV_0066|M.avium_104 RPEMVLAVVPHGTPKQVAKIVKSYVDAGLRVPKILDYG-AMAGLSHAQSS
MSMEG_6454|M.smegmatis_MC2_155 LEPLAQAIRDSGQLTAA---------------------------------
MUL_3202|M.ulcerans_Agy99 VDTFLFTIPDVARSDLIHTVGQEVLPAFD---------------------
MAB_0858|M.abscessus_ATCC_1997 LGYAICNFPEAAYDRSGIDLFVREVIGV----------------------
TH_2487|M.thermoresistible__bu VRELVTAQR-----------------------------------------
Mvan_2018|M.vanbaalenii_PYR-1 IPDEWLAPSATGSPQQCADRVRQELDYGADAVIMHGATPDELEPIVAAYR
Mb2975c|M.bovis_AF2122/97 VLPHAKVLRGLKKL-----
Rv2951c|M.tuberculosis_H37Rv VLPHAKVLRGLKKL-----
MLBr_00131|M.leprae_Br4923 LLPHARVLRGLKKL-----
MMAR_1758|M.marinum_M VLPHAKVLRGLKKL-----
Mflv_3380|M.gilvum_PYR-GCK TLPYNRVLRGLKKL-----
MAV_0066|M.avium_104 AANVRATEDELMRLCGDGS
MSMEG_6454|M.smegmatis_MC2_155 -------------------
MUL_3202|M.ulcerans_Agy99 -------------------
MAB_0858|M.abscessus_ATCC_1997 -------------------
TH_2487|M.thermoresistible__bu -------------------
Mvan_2018|M.vanbaalenii_PYR-1 AGRS---------------