For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDPLNRRQFLALAAAAAGVTAGCAGMGGGGSVKSGSGPIDFWSSHPGQSSAAERELIGRFQDRFPTLSVK LIDAGKDYDEVAQKFNAALIGTDVPDVVLLDDRWWFHFALIGVLTALDDLFGQVGVDTTDYVDSLLADYE FNGRHYAVPYARSTPLFYYNKAAWQQAGLPDRGLQSWSEFDEWGPELQRVVGAGRSAHGWANADLISWTF QGPNWAFGGAYSDKWTLTLTEPATIAAGNFYRNSIHGKGYAAVANDIANEFATGILASAVASTGSLPGIT ASARFDFGAAPLPTGPDAAPACPTGGAGLAIPAKLSEERKVNALKFIAFVTNPTNTAYFSQQTGYLPVRK SAVDDASERHYLADNPRARVALDQLPHTRTQDYARVFLPGGDRIISAGLESIGLRGADVTKTFTNIQKRL QVILDRQIMRKLAGHG
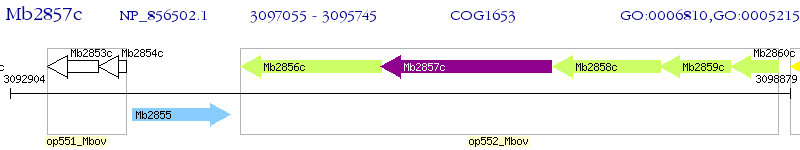
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2857c | ugpB | - | 100% (436) | sn-glycerol-3-phosphate-binding lipoprotein UGPB |
| M. bovis AF2122 / 97 | Mb2067c | - | 7e-19 | 25.74% (373) | sugar-binding lipoprotein |
| M. bovis AF2122 / 97 | Mb2345 | uspC | 5e-06 | 20.54% (443) | periplasmic sugar-binding lipoprotein UspC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2833c | ugpB | 0.0 | 99.31% (436) | sn-glycerol-3-phosphate-binding lipoprotein UGPB |
| M. leprae Br4923 | MLBr_01427 | - | 3e-20 | 26.88% (372) | putative ABC-transport lipoprotein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1900 | - | 0.0 | 75.51% (437) | ABC-type sugar transport protein |
| M. avium 104 | MAV_2476 | - | 3e-19 | 27.94% (383) | sugar ABC transporter, sugar-binding protein |
| M. smegmatis MC2 155 | MSMEG_0515 | - | 1e-14 | 22.56% (390) | sugar transporter sugar binding lipoprotein |
| M. thermoresistible (build 8) | TH_2011 | - | 8e-15 | 24.52% (363) | substrate binding protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2857c|M.bovis_AF2122/97 -MDPLNRRQFLALA-AAAAGVTAGCAGMGGGGSVKSGSGPIDFWSSHPGQ
Rv2833c|M.tuberculosis_H37Rv -MDPLNRRQFLALA-AAAAGVTAGCAGMGGGGSVKSGSGPIDFWSSHPGQ
MMAR_1900|M.marinum_M -MNSMHRRRFLSLASAAAAGVTAGCAGITNNVSIKSGPGPISFWSNHPGQ
MLBr_01427|M.leprae_Br4923 MHGKLFGRRSLLRG---AGALTAAALAPGAVGCSSDDD-ALTFFFAANPE
MAV_2476|M.avium_104 MLDRPFGRRSLLRG---AGALSAAALAPWSAGCGSDDDGALTFFFAANPE
MSMEG_0515|M.smegmatis_MC2_155 -----MIRRWLCLA---VVTAVACLLTACGGGSSSSGPVEIAVWHGYQDT
TH_2011|M.thermoresistible__bu ------VRRTPIRAVLAAVAAVCLTLTGCAGAGTLGSSGNTVTIALVSNS
*: . . . ..
Mb2857c|M.bovis_AF2122/97 SSAAERELIGRFQDRFPTLSVKLIDAGKDYDEVAQKFNAALIGTDVPDVV
Rv2833c|M.tuberculosis_H37Rv SSAAERELIGRFQDRFPTLSVKLIDAGKDYDEVAQKFNAALIGTDVPDVV
MMAR_1900|M.marinum_M SSGVEKVLIDRFQQQFPGLSVKLVDAGMDYDEVAEKFNAALIGDDVPDVV
MLBr_01427|M.leprae_Br4923 ETNARMRIVGEFQRDHPDIKVRAVLSGPG---VMQQLSTFCAGGKCPDVL
MAV_2476|M.avium_104 ERDARMRIIDEFARRHPDIKVRAVLSGPG---VMQQLSTFCVGGRCPDVL
MSMEG_0515|M.smegmatis_MC2_155 EGEAFKGLIDQYNKEHPDVHVTDLYSSND--LVLQKVLTAVRGGSAPDVA
TH_2011|M.thermoresistible__bu QMSDAQRLAPHFEAANPDIRLKFVTLSEN-QARAKITASTAMGGGEFDVV
. : .: * : : : . . : : * **
Mb2857c|M.bovis_AF2122/97 LLDDRWWFHFALIGVLTALDDLFG----QVGVDTTDYVDSLLADYEFNGR
Rv2833c|M.tuberculosis_H37Rv LLDDRWWFHFALSGVLTALDDLFG----QVGVDTTDYVDSLLADYEFNGR
MMAR_1900|M.marinum_M VLDDIWWFHFALSGVISPLDKLFR----QIGVDTSDYVDTLLADYEFNGR
MLBr_01427|M.leprae_Br4923 MAWDLTYAELADRGVLLDLNTLLGQDKAFAAELKSDSIEPLYETFTFNGG
MAV_2476|M.avium_104 MAWELSYAELADRGVLLDLGPLLARDKAFAQQLQADSIPALYETFTFNGK
MSMEG_0515|M.smegmatis_MC2_155 YMFGSWSPNIAKIPQVVDMSDVVS----QSDWNWDDFYPAEREAATVGDK
TH_2011|M.thermoresistible__bu MISNYETPQWAADGWLVNLSDYAR---QTPGYDEDDFIPSIRESLSYEGD
. * : :. * . .
Mb2857c|M.bovis_AF2122/97 HYAVPYARSTPLFYYNKAAWQQAGLPD-----RGLQSWSEFDEWGPELQR
Rv2833c|M.tuberculosis_H37Rv HYAVPYARSTPLFYYNKAAWQQAGLPD-----RGPQSWSEFDEWGPELQR
MMAR_1900|M.marinum_M HYALPYARSTPLFYYNKAVWQRAGLPD-----RGPNSWQEFDDWGPRLQR
MLBr_01427|M.leprae_Br4923 QYAFPEQWSGNYLFYNKQLFTNAGVQPPPCTWEQPWSFTEFLDTARALTK
MAV_2476|M.avium_104 QYALPEQWSGNYLFYNKRLFDEAGVPSPPTAWERPWDFSEFLDTARALTK
MSMEG_0515|M.smegmatis_MC2_155 IVGIPALVDNLAIVYNKKLFADAGIAPP----TADWTWDDFRAAAAKLTD
TH_2011|M.thermoresistible__bu MYAVPFYGESSFLMYRKDLFEKSGIDMP-----HNPTWQQVADAAAALDT
..* . : *.* : :*: : :. . *
Mb2857c|M.bovis_AF2122/97 VVGAGRSAHG-----WANADLISWTFQGPNWAFGGAYSDKWT--LTLTEP
Rv2833c|M.tuberculosis_H37Rv VVGAGRSAHG-----WANADLISWTFQGPNWAFGGAYSDKWT--LTLTEP
MMAR_1900|M.marinum_M VVDERQWAHG-----WANAELISWTFQGVNWTFDGSYSDLWT--LRFTDP
MLBr_01427|M.leprae_Br4923 RDSSGRVTQWGFVNTWLSYYTAGLFALNNGVPWSNPRMNPTH--LNFDDD
MAV_2476|M.avium_104 RDASGRAAQYGFVNTWGSYYSAGLFAMNNGVPWSDPRLNPTH--FNFDNA
MSMEG_0515|M.smegmatis_MC2_155 PAKG----QYGWLIPADGSEDTVWHYVPMLWEAGGDILTPDNEKAAFNSE
TH_2011|M.thermoresistible__bu PDMAGICLRG-----KPGWGEVLAPLDTVINTFGGRWYDMDWN-AQLDSP
: . .. : .
Mb2857c|M.bovis_AF2122/97 ATIAAGNFYRNSIHGKGYAAVAN-----DIANEFATGILASAVASTGSLP
Rv2833c|M.tuberculosis_H37Rv ATIAAGNFYRNSIHGKGYAAVAN-----DIANEFATGILASAVASTGSLA
MMAR_1900|M.marinum_M DTIAAGNFYRDSIHTKRYAAVAN-----NIANEFATGIMASAVASTGSLT
MLBr_01427|M.leprae_Br4923 AFIEAVQFYCDLTNKYQVAPDASEQQWMATADLFSLGKAAIALGGHWRYQ
MAV_2476|M.avium_104 AFQEAVQFYADLANKYRVAPNASETQSMSTPNLFAVGRAAMALGGHWRYQ
MSMEG_0515|M.smegmatis_MC2_155 AGVTALTMLQDMAVTDKSLYLDTTNE--NGPKLMNSGKVGMLITGPWDLS
TH_2011|M.thermoresistible__bu EVEEAVSFYVNLVRDHGEPGPATSGFG-ECATQFAQGRAAMWYDATSAVS
* : : . . : * . .
Mb2857c|M.bovis_AF2122/97 GITASARFDFGAAPLPTGPDAA-------PACP-TGGAGLAIPAKLSEER
Rv2833c|M.tuberculosis_H37Rv GITASARFDFGAAPLPTGPDAA-------PACP-TGGAGLAIPAKLSEER
MMAR_1900|M.marinum_M GITQMAKFDFGAAPVPTGPGGA-------PGCP-TGGAGLAIPTKLSDER
MLBr_01427|M.leprae_Br4923 TFMRAEGLDFDVTSLPIGPSAGTVPATRSGACSDIGATGLAIAA--SSSR
MAV_2476|M.avium_104 TYLRAEGLDFDVAPLPVGPAVG----KGQPACSDIGATGLAISS--SSPR
MSMEG_0515|M.smegmatis_MC2_155 ---QLSDIDYGVQVMPTFAGSS-------GAHQTISGPDNWVVFDNGDKR
TH_2011|M.thermoresistible__bu VLEDPAQSNVVGKVGYALAPTV-------VKADAGWLYTWALGIPASSDN
: . : .. .
Mb2857c|M.bovis_AF2122/97 KVNALKFIAFVTNPTNTAYFSQQTGYLPVRKS-AVDDASERHYLADNP-R
Rv2833c|M.tuberculosis_H37Rv KVNALKFIAFVTNPTNTAYFSQQTGYLPVRKS-AVDDASERHYLADNP-R
MMAR_1900|M.marinum_M KRNALRFIEYITNPVNTAYFSQRTGYLAVRKS-AADVPSEQKYLADHP-R
MLBr_01427|M.leprae_Br4923 KEQAWEFVKFATGPAGQALIGESCLFVPVLQS-AIYSTG----FAKAH-N
MAV_2476|M.avium_104 KEQAWEFVKFATGPVGQALIGESCLFVPVLRS-ALKSDG----FARAH-R
MSMEG_0515|M.smegmatis_MC2_155 KQASIDFVKWLTAPEQVKAFSLQTGDLPTRSS-VGDDQAVRDQLDQKLPG
TH_2011|M.thermoresistible__bu KDSAWKFISWMTDKNYIRLVGEELGWARVPPGNRLSTYQIPEYQQASEAF
* : *: : * .. . .
Mb2857c|M.bovis_AF2122/97 ARVALDQLPHTRTQDYARVFLP----------GGDRIISAGLESIG--LR
Rv2833c|M.tuberculosis_H37Rv ARVALDQLPHTRTQDYARVFLP----------GGDRIISAGLESIG--LR
MMAR_1900|M.marinum_M ARVAINQLPHTRPQDYARVFLP----------GADRIISAGLESIG--LR
MLBr_01427|M.leprae_Br4923 RVANLAVLTGGPVHSAGLPITP----------AWEKINALMDRNFGPVLR
MAV_2476|M.avium_104 RVGNLGVLTDGPAFSQGLPITP----------AWEKVNALMDRNFGPILR
MSMEG_0515|M.smegmatis_MC2_155 SSVFVENLNNAKKARPAVEQYP----------AIS--EALGQAIVAVMLG
TH_2011|M.thermoresistible__bu GQVTLESLENADTQHPTVQPVPYTGVQFLAIPEFQDLGTRVSQQISAAIA
: * * . : .. :
Mb2857c|M.bovis_AF2122/97 GADVTKTFTNIQKRLQVILDRQIMRKLAGHG
Rv2833c|M.tuberculosis_H37Rv GADVTKTFTNIQKRLQVILDRQIMRKLAGHG
MMAR_1900|M.marinum_M GTNVAKTFASIERQLQIILDRQIVRKLRQHG
MLBr_01427|M.leprae_Br4923 GVRPATSLAGLARAVDEVLNSP---------
MAV_2476|M.avium_104 GSRPATSLAGLSRSVDEVLRSP---------
MSMEG_0515|M.smegmatis_MC2_155 KEQPAAALNSAAEAADSALAGK---------
TH_2011|M.thermoresistible__bu GQKSVKDALAQAQKYAQVVGKTYQEKP----
. . :