For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRAKREAPKSRSSDRRRRADSPAAATRRTTTNSAPSRRIRSRAGKTSAPGRQARVSRPGPQTSPMLSPFD RPAPAKNTSQAKARAKARKAKAPKLVRPTPMERLAARLTSIDLRPRTLANKVPFVVLVIGSLGVGLGLTL WLSTDAAERSYQLSNARERTRMLQQHKEALERDVREAASAPALAEAARRQGMIPTRDTAHLAQDPDGNWV VVGTPKPADGVPPPPLNTKLPEDPPPPPKPAAVPLEVPVRVTPGPDDPAPPARSGPEVLVRTPDGTATLG GATHLPTQAGPQLPGPVPIPGAPGPMPAPPLGAVPSPAPAENPVPLQVGAAPPAGLPGPAPVAATPGLSG GSQPMVAPPAPVPANGEQFGPVTAPVPTAPGAPR
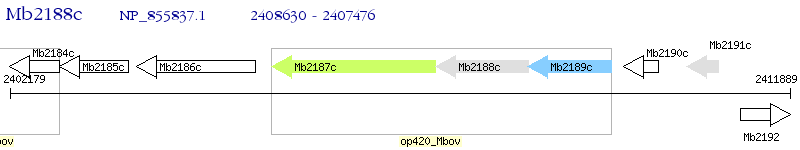
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2188c | - | - | 100% (384) | hypothetical protein Mb2188c |
| M. bovis AF2122 / 97 | Mb0177 | mce1C | 7e-13 | 36.02% (161) | MCE-family protein MCE1C |
| M. bovis AF2122 / 97 | Mb3906 | - | 1e-11 | 29.47% (207) | hypothetical protein Mb3906 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2981 | - | 8e-83 | 49.74% (382) | hypothetical protein Mflv_2981 |
| M. tuberculosis H37Rv | Rv2164c | - | 0.0 | 99.74% (384) | hypothetical protein Rv2164c |
| M. leprae Br4923 | MLBr_00907 | - | 1e-124 | 61.71% (397) | hypothetical protein MLBr_00907 |
| M. abscessus ATCC 19977 | MAB_1999 | - | 6e-71 | 45.14% (370) | hypothetical protein MAB_1999 |
| M. marinum M | MMAR_3201 | - | 1e-127 | 60.79% (431) | proline rich membrane protein |
| M. avium 104 | MAV_2329 | - | 1e-119 | 58.24% (431) | hypothetical protein MAV_2329 |
| M. smegmatis MC2 155 | MSMEG_4234 | - | 6e-88 | 50.50% (400) | hypothetical protein MSMEG_4234 |
| M. thermoresistible (build 8) | TH_3807 | - | 5e-09 | 32.26% (155) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_3509 | - | 1e-126 | 59.91% (429) | proline rich membrane protein |
| M. vanbaalenii PYR-1 | Mvan_3530 | - | 3e-84 | 51.78% (365) | FHA domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2981|M.gilvum_PYR-GCK MKAKKPLRNAAAEPARRSERTSGRSRATTTKPKAGGKRGTAREAKRTPVD
Mvan_3530|M.vanbaalenii_PYR-1 MKAQKPMRKTATG-------EPGRARTSAKKSPRAAKRRPVRETKRTPVD
MSMEG_4234|M.smegmatis_MC2_155 MKAKR----------------PGPVRDADRRR---GKRQPG--VARGSAE
MAB_1999|M.abscessus_ATCC_1997 MIGVR-----------------KKAGEAPKKT------KPAKSAKPTKRR
Mb2188c|M.bovis_AF2122/97 MRAKREAPKSR----------SSDRRRRADSPAAATRRTTTNSAPSRRIR
Rv2164c|M.tuberculosis_H37Rv MRAKREAPKSR----------SSDRRRRADSPAAATRRTTTNSAPSRRIR
MMAR_3201|M.marinum_M MKAKREARVPG----------KRQSGDRGGGRDGSVARTTRRTPADASRR
MUL_3509|M.ulcerans_Agy99 MKAKREARVPG----------KRQSGDRGGGRDGSVARTTRRTPADASRR
MLBr_00907|M.leprae_Br4923 MKAQRDTPIRR----------GDSGRPGGRDGAARSGKRTANEAGSRRLR
MAV_2329|M.avium_104 MKAERETAKRR----------GGSDRRGARDGSARRGRRPAADAGAAR-R
TH_3807|M.thermoresistible__bu -------------------------------------MRATRKLITSSVI
.
Mflv_2981|M.gilvum_PYR-GCK AQPRGRKTTRD-ARPQRSAPRTTPISRPVDAPARPKSAT---QAKARAKA
Mvan_3530|M.vanbaalenii_PYR-1 APPRGRQQTRD-ARPQRSAPRTTPIARPKDVPARPKSTS---QAKARAKA
MSMEG_4234|M.smegmatis_MC2_155 APPRKRRPSREPQRPLRSAPQTGPIPRQKTEPARPKTAS---QAKARAKA
MAB_1999|M.abscessus_ATCC_1997 NGASKATATGPSARPRRSAPQTMPIPVQRQAPEKIRPATSTRQAKARAKA
Mb2188c|M.bovis_AF2122/97 SRAGKTSAPGRQARVSRPGPQTSPMLSPFDRPAPAKNTS---QAKARAKA
Rv2164c|M.tuberculosis_H37Rv SRAGKTSAPGRQARVSRPGPQTSPMLSPFDRPAPAKNTS---QAKARAKA
MMAR_3201|M.marinum_M VRAGKKKAPNREARIPMSGPQTTPMPRPAERPARPKNTG---QAKARAKA
MUL_3509|M.ulcerans_Agy99 GRAGKKKAPNREARIPMSGPQTTPMPRPAERPARPKNTG---QAKARAKA
MLBr_00907|M.leprae_Br4923 THAGKISASAREVGVPKSGPRTSPMSRPVERPARPRNTT---QAKARAKA
MAV_2329|M.avium_104 ARGGPASAPTRESRAPKPGPQTSPVARPAERSARPKNTS---QAKARAKA
TH_3807|M.thermoresistible__bu AAGGAVAVALTCSATAAAQPAPAPAPSP-GVPFMDQLAT-----------
. . * . * . : :
Mflv_2981|M.gilvum_PYR-GCK RKAKAPKVVRPPLRERLLVRLASIDLNPRRLAARVPFVVLIISALGLGLG
Mvan_3530|M.vanbaalenii_PYR-1 RKAKAPKVVRPPLRQRILVRLASIELDPRRLIARVPFVVLVIGALGLGLG
MSMEG_4234|M.smegmatis_MC2_155 RKAKAPKVVRPPLRERILLRLSSIELNPRELVARVPFVVLVIAALGLGLG
MAB_1999|M.abscessus_ATCC_1997 RKAKAPKVIRVPLRERVLARLSEVDLRPHVWITRVPFVVFVIGALGVGLA
Mb2188c|M.bovis_AF2122/97 RKAKAPKLVRPTPMERLAARLTSIDLRPRTLANKVPFVVLVIGSLGVGLG
Rv2164c|M.tuberculosis_H37Rv RKAKAPKLVRPTPMERLAARLTSIDLRPRTLANKVPFVVLVIGSLGVGLG
MMAR_3201|M.marinum_M RKAKAPKVVRPKLTERMAVRLASMDLRPHALLSRVPFVVLVIGSLGVGLG
MUL_3509|M.ulcerans_Agy99 RKAKAPKVVRPKLTERMAVRLASMDLRPHALLSRVPFVVLVIGSLGVGLG
MLBr_00907|M.leprae_Br4923 RKAKAPKVVRPRLGECLIARLALIDLRPRTLVNKVPFVVLVISSLGVGLG
MAV_2329|M.avium_104 RKAKAPKVVRPRLTERLAARLAAIDLRPRTLASKVPFVVLVIGALGVGLG
TH_3807|M.thermoresistible__bu AAANAPAILQS----------AAAALGGTNTMPSAPVVAPPPAAT-----
*:** ::: : * .*.*. .:
Mflv_2981|M.gilvum_PYR-GCK VTLWLSTDAAERSYQLGSARQLNQALMQQKEALERDVLEAQAAPALAEAA
Mvan_3530|M.vanbaalenii_PYR-1 VTLWLSTDAAERSYQLGAARQLNQALLQQKEGLERDVLEAQAAPALAEAA
MSMEG_4234|M.smegmatis_MC2_155 ITLWLSTSAAERSYQLGHARQVNQGLLQQKEALERDVLEAQAAPALAEAA
MAB_1999|M.abscessus_ATCC_1997 ITLWLSTDAAERSYQLGTQRRTNEELLQRKEALERDVLRAESAPALADSA
Mb2188c|M.bovis_AF2122/97 LTLWLSTDAAERSYQLSNARERTRMLQQHKEALERDVREAASAPALAEAA
Rv2164c|M.tuberculosis_H37Rv LTLWLSTDAAERSYQLSNARERTRMLQQHKEALERDVREAASAPALAEAA
MMAR_3201|M.marinum_M LTLWLSTDAAERSYQLSHARERTRMLQQQKESLERDVREAESAPALAEAA
MUL_3509|M.ulcerans_Agy99 LTLWLSTDAAERSYQLSHARERTRMLQQQKESLERDVREAESAPALAEAA
MLBr_00907|M.leprae_Br4923 LTLWLSTDSAERSYQLGDAREQARMLQQQKEALERDVREAESAPALAETA
MAV_2329|M.avium_104 LTLWLSTDSAERSYKLSHARERTRLLQQQKEALERDVREAEAAPALAEAA
TH_3807|M.thermoresistible__bu ----AQITVPQPALPATVAPATAPIIKQP-----GTVTPTAPGVPLPTAE
. .: : : * * : .. .*. :
Mflv_2981|M.gilvum_PYR-GCK RELGMIPSRDTAHLVQDASGNWVVVGTPKPAEGVPPPPLNAPLP---EPA
Mvan_3530|M.vanbaalenii_PYR-1 RNLGMIPSRDTAHLVQDPSGNWVVVGTPKPAEGVPPPPLNAPLP---EPA
MSMEG_4234|M.smegmatis_MC2_155 RNLGMIPSRDTAHLVQDPTGNWVVVGTPKPAEGVPPPPLNAPLPPDAPPP
MAB_1999|M.abscessus_ATCC_1997 RDLGMIPSRDIAHLVRDPQGNWLLVGAPKPAEGVAPAPLNSVIP------
Mb2188c|M.bovis_AF2122/97 RRQGMIPTRDTAHLAQDPDGNWVVVGTPKPADGVPPPPLNTKLP---ED-
Rv2164c|M.tuberculosis_H37Rv RRQGMIPTRDTAHLVQDPDGNWVVVGTPKPADGVPPPPLNTKLP---ED-
MMAR_3201|M.marinum_M RKQGMIPTRDTAHLVQAPDGNWVVVGTPKPADGVPPPPLNTKLP---EEG
MUL_3509|M.ulcerans_Agy99 RKQGMIPTRDTAHLVQAPDGNWVVVGTPKPADGVPPPPLNSKLP---EEG
MLBr_00907|M.leprae_Br4923 RKQGMIPTRDTAHLVQGPGGNWVVVGTPKPADGVPPPPLNTKLP---DAG
MAV_2329|M.avium_104 RKQGMIPTRDTAHLVQDPSGNWVVVGTPKPADGVPPPPLNSKLP---DEA
TH_3807|M.thermoresistible__bu ATLPNVAGTPVPQQVSLPAD----VSALLPAAGSQLANLMPAVP--NEPG
:. .: . . . *.: ** * . * . :*
Mflv_2981|M.gilvum_PYR-GCK PPAPPAPRVVEPREVPVRIPSAA---------TPLAVPGMVPGA------
Mvan_3530|M.vanbaalenii_PYR-1 PPAPPAPRVIEPLEVPVRLPSVT---------APQAIPGTAPAL------
MSMEG_4234|M.smegmatis_MC2_155 PPAPPAPRVVEPREVPIRVPEAPGLQGLPGIQAAPGIPGAVQGV------
MAB_1999|M.abscessus_ATCC_1997 DDADAVPKAGNSVEVPVQLPPVP--------PPVAVVPHMAP--------
Mb2188c|M.bovis_AF2122/97 PPPPPKPAAV-PLEVPVRVTPGPDDPAPPARSGPEVLVRTPDGTATLGGA
Rv2164c|M.tuberculosis_H37Rv PPPPPKPAAV-PLEVPVRVTPGPDDPAPPARSGPEVLVRTPDGTATLGGA
MMAR_3201|M.marinum_M PPAPPKPAAL-PPEVPVRVMPGPDDPALLPRTGPQLLPRTPDGSSTLGAG
MUL_3509|M.ulcerans_Agy99 PPAPPKPAAL-PPEVPVRVMPGPDDPALLPRTGPQLLPRTPDGSSTLGAD
MLBr_00907|M.leprae_Br4923 PPSL-KPPEI-PLEVPVRVVPGPGGPPPPARSGPQMWLRVPDGATTLGGQ
MAV_2329|M.avium_104 PQPP-KPAPP-AAEVPIRVELAPGGP-VPARSGPEALLRAPDGASTLGGQ
TH_3807|M.thermoresistible__bu APAPATPPMLPSGLFPTSALP-----------------------------
. * . .*
Mflv_2981|M.gilvum_PYR-GCK -----------------------PAPLPGL--------------------
Mvan_3530|M.vanbaalenii_PYR-1 -----------------------PLAVPGA--------------------
MSMEG_4234|M.smegmatis_MC2_155 -----------------------PGAVPGE--------------------
MAB_1999|M.abscessus_ATCC_1997 -----------------------PTAAP----------------------
Mb2188c|M.bovis_AF2122/97 T---HLPTQAGPQLP-------GPVPIPG---------------------
Rv2164c|M.tuberculosis_H37Rv T---HLPTQAGPQLP-------GPVPIPG---------------------
MMAR_3201|M.marinum_M TN--HLPGQPPIEIPGQVPGAPGPVPLPGQ-----------LPGVAGPVT
MUL_3509|M.ulcerans_Agy99 TN--HLPGQPPIEIPGQVPGAPGPVPLPGQ-----------LPGVAGPVT
MLBr_00907|M.leprae_Br4923 HLPQELPQLP----GMLNGPAAAQVQVPG-----------FMPT------
MAV_2329|M.avium_104 HLAPAVPPVPGTPPGVPGGPVAAPPALPGGPVAAVPPASPAMPTGPATGR
TH_3807|M.thermoresistible__bu --------------------------------------------------
Mflv_2981|M.gilvum_PYR-GCK ----AP---VPGSE----------------------------VPHAVSAD
Mvan_3530|M.vanbaalenii_PYR-1 ----VPNVAVPNVE----------------------------VPHAVSGQ
MSMEG_4234|M.smegmatis_MC2_155 ----TP-HVAPGAQPQ--------------------------APGPVIGP
MAB_1999|M.abscessus_ATCC_1997 ----APVVEAPIGQ----------------------------DPAAVPQV
Mb2188c|M.bovis_AF2122/97 APGPMPAPPLGAVPSP-------------APAENPVPL---QVGAAPPAG
Rv2164c|M.tuberculosis_H37Rv APGPMPAPPLGAVPSP-------------APAENPVPL---QVGAAPPAG
MMAR_3201|M.marinum_M GPGQLPGPAAGAVQVPPAPVVLPAPVPAQVPAEVPVPVPVPAVTAPPVAV
MUL_3509|M.ulcerans_Agy99 GPGQLPGPAAGAVQVPPAPVVLPAPVPAQVPAEVPVPV--PAVTAPPVAV
MLBr_00907|M.leprae_Br4923 -----PGLPIPGSTMR--------------------------VPVPAP--
MAV_2329|M.avium_104 PPGLTPGLPMPGLPMPAGPAVAP------APAEVSIPL--GGLPVPAPGQ
TH_3807|M.thermoresistible__bu --------------------------------------------------
Mflv_2981|M.gilvum_PYR-GCK IPMP--GPV-LPAAAPDPALVAP------------APAPAPAPAPAPVPG
Mvan_3530|M.vanbaalenii_PYR-1 LPAP--GPVPVPVPGPAPVPAAP------------LDAGTSQLPPAPAPG
MSMEG_4234|M.smegmatis_MC2_155 LTGP--APMAVPMPVPAEVPLATPQAPQQAAPLVNTPAPQQVPAPAPLPG
MAB_1999|M.abscessus_ATCC_1997 APVPA-VPADVPPPVADAPQASN-----------ITNSPVTGEQFNPVAT
Mb2188c|M.bovis_AF2122/97 LPGP------APVAATPGLSGGSQ---PMVAP--PAPVPANGEQFGPVTA
Rv2164c|M.tuberculosis_H37Rv LPGP------APVAATPGLSGGSQ---PMVAP--PAPVPANGEQFGPVTA
MMAR_3201|M.marinum_M VPAAPQAPLAAPVPGTAGLPVAPQAGVPAPVP--ALPAPASGVQPGPAAA
MUL_3509|M.ulcerans_Agy99 VPVAPQTPLAAPVPGTAGLPVAPQAAVPAPVP--ALPAPASGIQPGPAAA
MLBr_00907|M.leprae_Br4923 ------APTEVPVRLQPGLVSPAVTSPVISTS--PVPTPVNSEQFGPVTA
MAV_2329|M.avium_104 LPAP--LPPEVPVPVQLGRPG-AAPQPVVPAA--PLPQPVNGAQPGAAPV
TH_3807|M.thermoresistible__bu --------------------------------------------------
Mflv_2981|M.gilvum_PYR-GCK ---AGA---------------------------------
Mvan_3530|M.vanbaalenii_PYR-1 ---APA---------------------------------
MSMEG_4234|M.smegmatis_MC2_155 ---AVPQGAPLGALPGAQPAVPDPATAVEHFSPAVAPVA
MAB_1999|M.abscessus_ATCC_1997 ---TSPLPAA-----------------------------
Mb2188c|M.bovis_AF2122/97 PVPTAPGAPR-----------------------------
Rv2164c|M.tuberculosis_H37Rv PVPTAPGAPR-----------------------------
MMAR_3201|M.marinum_M ---AAPGVSR-----------------------------
MUL_3509|M.ulcerans_Agy99 ---AAPGVSR-----------------------------
MLBr_00907|M.leprae_Br4923 ---TAPGTSR-----------------------------
MAV_2329|M.avium_104 ---ASPGAPR-----------------------------
TH_3807|M.thermoresistible__bu ---------------------------------------