For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RYVVTGGTGFIGRRVVSEILVRDASAEVWVLVRRESLNRFEHLALDWGERAKPLVGDLRADDLGLTGADV ADLGDVTHVVHCAAIYDITVDEAAQRAANVDGTRAVIALARRLEATLHHVSSIAVAGNHRGVFTEDDFDV AQDLPTPYHQTKFEAEMLVRATTDVPHRIYRPAVVVGDSRTGEMDKIDGPYYFFGVLARLAALPRFTPMA LPDTGRTNVVPVDFVAKALVHLMHVEGADGQTFHLTAPNTVGLVDIYRGVAGAAGLPPLRAKLPRVTAAP VLDARGRARIVRNMVATQLGIPGEVFDVAELRPTFTADNTDTALRGTGIAVPEFASYAPQLFRYWAEHLD PDRARRCDPAGPLVGRHVIITGASSGIGRASAIAVARRGAVVFALARNGEALDELIAEIRADGGQAHAFT CDVTDSASVEHTVKDILGRFDHVDYLVNNAGRSIRRSVANSTDRLHDYERVMAVNYFGAVRMVLALLPHW RERRFGHVVNVSSAGVQAHTPKYSAYLPSKAALDAFSDVVSTETLSDHITFTNIHMPLVKTPMIAPSRRL NPVPPISAEHAAAMVVRALVEKPPRIDTPLGTFADFGNYLTPRVARRFLHQLYLGYPDSAAARGLAPAEP RPASTAGPGLPERRPKRPARSRRGVRVPRAVGRPVKRAVRTLPGVHW*
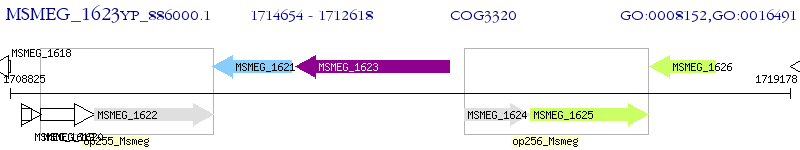
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1623 | - | - | 100% (678) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1073 | - | 1e-48 | 40.73% (275) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6859 | - | 3e-23 | 35.32% (218) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3423 | acrA1 | 0.0 | 72.71% (678) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4896 | - | 0.0 | 75.07% (678) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv3391 | acrA1 | 0.0 | 72.71% (678) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 1e-22 | 37.50% (192) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3710 | - | 0.0 | 67.70% (678) | short chain dehydrogenase |
| M. marinum M | MMAR_1153 | acrA1 | 0.0 | 73.42% (681) | short-chain dehydrogenase, AcrA1 |
| M. avium 104 | MAV_4340 | - | 0.0 | 75.84% (683) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3707 | - | 1e-28 | 39.38% (193) | PUTATIVE oxidoreductase, short-chain dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_0918 | acrA1 | 0.0 | 73.42% (681) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1527 | - | 0.0 | 76.13% (662) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4896|M.gilvum_PYR-GCK ----------MRYVVTGGTGFIGSRLIGRLLARPDVTAVHVLVRRGSLGR
Mvan_1527|M.vanbaalenii_PYR-1 --------------------------MTRLLSDPGTAAVHVLVRRDSLGR
MSMEG_1623|M.smegmatis_MC2_155 ----------MRYVVTGGTGFIGRRVVSEILVRDASAEVWVLVRRESLNR
Mb3423|M.bovis_AF2122/97 ----------MRYVVTGGTGFIGRHVVSRLLDGRPEARLWALVRRQSLSR
Rv3391|M.tuberculosis_H37Rv ----------MRYVVTGGTGFIGRHVVSRLLDGRPEARLWALVRRQSLSR
MAV_4340|M.avium_104 MRGRHRHDVFMRYVVTGGTGFIGRRVVTRLLQTRPDAQVWVLVRRQSLGR
MMAR_1153|M.marinum_M ----------MRYVVTGGTGFIGRRVVSRLLQSDADARVWVLVRRQSLSR
MUL_0918|M.ulcerans_Agy99 ----------MRYVVTGGTGFIGRRVVSRLLQSDADARVWVLVRRQSLSR
MAB_3710|M.abscessus_ATCC_1997 ----------MKYIVTGGTGFIGRRIVTRILETQPAAEVAILVRRESLSR
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 ----------------MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGP
Mflv_4896|M.gilvum_PYR-GCK FERLARD----WDDRVHALVGDLTDPDLGLTDATVAELGPVDHVVHCAAI
Mvan_1527|M.vanbaalenii_PYR-1 FERLALN----WDDRVQPLVGELTVPGLGTSDVTVAELGRIDHVVHCAAV
MSMEG_1623|M.smegmatis_MC2_155 FEHLALD----WGERAKPLVGDLRADDLGLTGADVADLGDVTHVVHCAAI
Mb3423|M.bovis_AF2122/97 FERLAGQ----WGDRVRPLVGDL--TELELSERTIAELGDIDHVLHCAAV
Rv3391|M.tuberculosis_H37Rv FERLAGQ----WGDRVRPLVGDL--TELELSERTIAELGDIDHVLHCAAV
MAV_4340|M.avium_104 FERLSAQGDTPWGDRVKPLVGEL--PALELSDETLAELGHVDHLVHCAAI
MMAR_1153|M.marinum_M FERLASD----WGDRVKPLVGEL--PELPVTEQISAELGQIDHVVHCAAV
MUL_0918|M.ulcerans_Agy99 FERLATD----WGDRVKPLVGEL--PELPVTEQISAELGQIDHVVHCAAV
MAB_3710|M.abscessus_ATCC_1997 FEKLAEQ----WDDRVQPLVGDLTQPDLGLPAE--GDPVTADHIVHCAAI
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 AIVLVHG----FPDSHVLWDGVVPLLAKRFRIVRYDNRG----VGQSSVP
Mflv_4896|M.gilvum_PYR-GCK YDMTAPENRQRAANVEGTRAVIALSRRLGATLHHVSSIAVAGTHPGVFTE
Mvan_1527|M.vanbaalenii_PYR-1 YDITAPEDRQRAANVDGTGAVVALSRRLGATLHHVSSIAVAGTYRGVFTE
MSMEG_1623|M.smegmatis_MC2_155 YDITVDEAAQRAANVDGTRAVIALARRLEATLHHVSSIAVAGNHRGVFTE
Mb3423|M.bovis_AF2122/97 HDTTW---------ADATRAVIELAARLDATFHHVSSIAVAGDFAGHYTE
Rv3391|M.tuberculosis_H37Rv HDTTW---------ADATRAVIELAARLDATFHHVSSIAVAGDFAGHYTE
MAV_4340|M.avium_104 YDITAGEAEQRAANVDGTRAVIELAQRLGATLHHVSSIAVAGDFPGEYTE
MMAR_1153|M.marinum_M YDITAGEDQQRAANVDGTRAVIELTRRLDATLHHVSSIAVAGDFAGEYTE
MUL_0918|M.ulcerans_Agy99 YDITAGEDQQRAANVDGTRAVIELTRRLDATLHHVSSIAVAGDFAGEYTE
MAB_3710|M.abscessus_ATCC_1997 YDITVDDKAQRAANVEGTRSVIALAKRTGAILHHISSIAVAGSYEGEFTE
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 KPVSAYTMTRFADDFAAVIDELSPDRPVHVLAHDWGSVGVWEYLSRSGAS
Mflv_4896|M.gilvum_PYR-GCK DDVDVGQELPTPYHRTKFEAEQLVRAADGLRFRIYRPAVVVGDSRTGEID
Mvan_1527|M.vanbaalenii_PYR-1 DDFDVAQDLPTPYHRTKFEAELLVRSAEGLRFRIYRPAVVVGDSRTGEMD
MSMEG_1623|M.smegmatis_MC2_155 DDFDVAQDLPTPYHQTKFEAEMLVRATTDVPHRIYRPAVVVGDSRTGEMD
Mb3423|M.bovis_AF2122/97 ADFDVGQRLPTPYHRMTFEAERLVRSTPGLRYRIYRPAVVVGDSRTGEMD
Rv3391|M.tuberculosis_H37Rv ADFDVGQRLPTPYHRMTFEAERLVRSTPGLRYRIYRPAVVVGDSRTGEMD
MAV_4340|M.avium_104 DDFDVGQQLPTPYHQTKFEAELLVRSAPGLRHRIYRPAVVVGDSRTGEMD
MMAR_1153|M.marinum_M ADFDVGQQLPSPYHRTKFEAELLVRSSTGLRYRIYRPAVVVGDSRTGEMD
MUL_0918|M.ulcerans_Agy99 ADFDVGQQLPSPYHRTKFEAELLVRSSTGLRYRIYRPAVVVGDSRTGEMD
MAB_3710|M.abscessus_ATCC_1997 DDFDVNQDLPTPYHQTKFEAELLVRSEPGLRYRIYRPAVVVGDSRTGEMD
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRVASFTSVSGPSQ------DQLVDYILSGLRRPWRPRTFARAVSQ----
Mflv_4896|M.gilvum_PYR-GCK KIDGPYYFFGVLSALAALPRFTPMMLPDVGRTNLVPVDFVVDALDVLMHA
Mvan_1527|M.vanbaalenii_PYR-1 KIDGPYYFFGVLSALAALPGFTPMLLPDVGRTNIVPVDFVVEAVAALMHV
MSMEG_1623|M.smegmatis_MC2_155 KIDGPYYFFGVLARLAALPRFTPMALPDTGRTNVVPVDFVAKALVHLMHV
Mb3423|M.bovis_AF2122/97 TIDGPYYLFGVLAKLAVLPSFTPMLLPDIGRTNIVPVDYVADALVALMHA
Rv3391|M.tuberculosis_H37Rv TIDGPYYLFGVLAKLAVLPSFTPMLLPDIGRTNIVPVDYVADALVALMHA
MAV_4340|M.avium_104 KVDGPYYFFGVLAKLAVLPRFTPMLLPDTGRTNIVPVDYVADALVALMHA
MMAR_1153|M.marinum_M KIDGPYYFFGVLARLAALPSLTPILLPDTGRTNIVPVDYVVDALVALIHE
MUL_0918|M.ulcerans_Agy99 KIDGLYYFFGVLARLAALPSLTPILLPDTGRTNIVPVDYVVDALVALIHE
MAB_3710|M.abscessus_ATCC_1997 KIDGPYYFFGVFAKLAKLPSFTPMMLPRAGRSNIVPVDYVVDATVELMHQ
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 ------LFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMVDNISTDQIHH
Mflv_4896|M.gilvum_PYR-GCK DGR---DGQTFHLTAPKNTGLRGIYRGVARAAGLPPLVGVLPRAAAAPLL
Mvan_1527|M.vanbaalenii_PYR-1 DGR---DGQTFHLTAPKTTGLRGIYRGIAAAAGLPPLVGSLPRATAAPLL
MSMEG_1623|M.smegmatis_MC2_155 EGA---DGQTFHLTAPNTVGLVDIYRGVAGAAGLPPLRAKLPRVTAAPVL
Mb3423|M.bovis_AF2122/97 DGR---DGQTFHLTAPTAIGLRGIYRGIAGAAGLPPLLGTLPGFVAAPVL
Rv3391|M.tuberculosis_H37Rv DGR---DGQTFHLTAPTAIGLRGIYRGIAGAAGLPPLLGTLPGFVAAPVL
MAV_4340|M.avium_104 DGL---DGQTFHLTAEKTIGLRGIYRGVAKAAGLPPLRGSLPGAVAAPVL
MMAR_1153|M.marinum_M DNMAFPNGRTFHLTAPQSVGLRGIYQAVAKEAGLPPARGALPGSVVTPVL
MUL_0918|M.ulcerans_Agy99 DNMAFPNGRTFHLTAPQSVGLRGIYQAVTKEAGLPPARGVLPGSVVTPVL
MAB_3710|M.abscessus_ATCC_1997 EGR---DGQAFHLTNPEVTYLRDIYSGIAAAAGLPPVRGSLPHAVATPFL
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 SAT--LARDAAHSVKTYPANYFRAFSGRRRGKGVPVVN-----VPVQLIV
Mflv_4896|M.gilvum_PYR-GCK QVSGRAKAIRNMAATQLGIPGEVLDVVELAPTFTSERTDEALCDSGISVP
Mvan_1527|M.vanbaalenii_PYR-1 HVTGRAKVLRNMVATQLGIPAEILDIVELAPTFTSEHTDEALRGTGISVP
MSMEG_1623|M.smegmatis_MC2_155 DARGRARIVRNMVATQLGIPGEVFDVAELRPTFTADNTDTALRGTGIAVP
Mb3423|M.bovis_AF2122/97 NARGRAKVLRNMAATQLGIPAEIFDVVGCAPTFTSDTTREALRGTGIHVP
Rv3391|M.tuberculosis_H37Rv NARGRAKVLRNMAATQLGIPAEIFDVVGCAPTFTSDTTREALRGTGIHVP
MAV_4340|M.avium_104 KVRGRARVVRDMAATQLGIPAQVFDLVDLAPTFVSEKTRNALAGTGIEVP
MMAR_1153|M.marinum_M KARGRAKVLRNMAATQLGIPSEIFDVLDLKPTFISDETQKALRGSGVRLP
MUL_0918|M.ulcerans_Agy99 KARGRAKVLRNMAATQLGIPSEIFDVLDLKPTFISDETQKALRGSGVRLP
MAB_3710|M.abscessus_ATCC_1997 RARGTLKVWRNMVVTQLGIPPEVIDITEIMPTFISESTQEALRPSGIKVP
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 NTMDRYVRPYGYDATARWVPRLWRRDIKAG-HFSPMSHPQVMAAAVHDLA
Mflv_4896|M.gilvum_PYR-GCK EFASYAPGLWRYWAEHLDPNRARQDDPAGPLVGRHVVITGASSGIGRASA
Mvan_1527|M.vanbaalenii_PYR-1 EFASYAPALWRYWAQHLDPDRARRDDPAGPLVGKHVIITGASSGIGRAAA
MSMEG_1623|M.smegmatis_MC2_155 EFASYAPQLFRYWAEHLDPDRARRCDPAGPLVGRHVIITGASSGIGRASA
Mb3423|M.bovis_AF2122/97 EFATYAPGLWRYWAEHLDPDRARRNDP---LLGRHVIITGASSGIGRASA
Rv3391|M.tuberculosis_H37Rv EFATYAPGLWRYWAEHLDPDRARRNDP---LLGRHVIITGASSGIGRASA
MAV_4340|M.avium_104 EFAGYAPRLWRYWAEHLDPDRARRDDPRGPLYGRHVIVTGASSGIGRASA
MMAR_1153|M.marinum_M DFAGYAPKLWRYWAQHLDPDRALRDDPRGPLVGRHVIITGASSGIGRASA
MUL_0918|M.ulcerans_Agy99 DFAGYAPKLWRYWAQHLDPDRALRDDPRGPLVGRHVIITGASSGIGRASA
MAB_3710|M.abscessus_ATCC_1997 PFGSYAGRLYQYWRRNLDPERYRRDDPAGPLVGRHVIITGASSGIGRAAA
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DLVDGKQPSRALLRAQVGRSRD-------TFGDTLVSVTGAGSGIGRETA
Mflv_4896|M.gilvum_PYR-GCK VAVAARGATVFALARNAEALDDLIAEIRESGGDAHAFTCDVTDPSAVEHT
Mvan_1527|M.vanbaalenii_PYR-1 IAVAARGATVFALARNAEALDDLIAEIRAAGGDAHAFTCDVTDSAAVEHT
MSMEG_1623|M.smegmatis_MC2_155 IAVARRGAVVFALARNGEALDELIAEIRADGGQAHAFTCDVTDSASVEHT
Mb3423|M.bovis_AF2122/97 IAVAKRGATVFALARNGNALDELVTEIRAHGGQAHAFTCDVTDSASVEHT
Rv3391|M.tuberculosis_H37Rv IAVAKRGATVFALARNGNALDELVTEIRAHGGQAHAFTCDVTDSASVEHT
MAV_4340|M.avium_104 IAIAQRGATVFALARNGAALDALVDEIRANGGAAHAFTCDVTDSASVEHT
MMAR_1153|M.marinum_M LAVAQRGGCVFALARNADALDQLVAEIRAGGGQAYAFTCDITDSTSVEHT
MUL_0918|M.ulcerans_Agy99 LAVAQRGGCVFALARNADALDQLVAEIRAGGGQAYAFTCDITDSTSVEHT
MAB_3710|M.abscessus_ATCC_1997 IAVARRGGTVFAVARDGEALDQLVAEIREDGGKAHAFPCDLTDYAAVDDT
TH_3707|M.thermoresistible__bu -------------VRADGRADQGLDAARX-GGTGRALACDLTDLDAIDEL
MLBr_00862|M.leprae_Br4923 LALARRGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAF
. ** . * . *::* :::
Mflv_4896|M.gilvum_PYR-GCK VKDILGRFGHVDYLVNNAGRSIRRSVTASTDRFHDYERVMAVNYFGAVRM
Mvan_1527|M.vanbaalenii_PYR-1 VKDILGRFDHVDYLVNNAGRSIRRSVAASTDRLHDYERVMAVNYFGSVRM
MSMEG_1623|M.smegmatis_MC2_155 VKDILGRFDHVDYLVNNAGRSIRRSVANSTDRLHDYERVMAVNYFGAVRM
Mb3423|M.bovis_AF2122/97 VKDILGRFDHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
Rv3391|M.tuberculosis_H37Rv VKDILGRFDHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
MAV_4340|M.avium_104 VKDILGRFDHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
MMAR_1153|M.marinum_M VKDILGRFGHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
MUL_0918|M.ulcerans_Agy99 VKDILGRFGHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
MAB_3710|M.abscessus_ATCC_1997 VKSILGQFGHVDYLVNNAGRSIRRSVVNTVDRFHDYERVMAINYFGAVRL
TH_3707|M.thermoresistible__bu VLKIEQDLGGVDILINNAGKSIRRPLAESLERWHDVERTMNLNYYAPLRL
MLBr_00862|M.leprae_Br4923 AEQVSAKHGVPDIVVNNAGIGQAGRFLDTPP--EEFDRVLAVNLGGVVNC
. .: . * ::**** . . : .: :*.: :* . :.
Mflv_4896|M.gilvum_PYR-GCK VLALLPHWRERRFG-HVVNVSSAGVQANNPRYSAYIPSKAALDAFSEVVG
Mvan_1527|M.vanbaalenii_PYR-1 VLALLPHWRERRFG-HVVNVSSAGVQANNPRYSAYIPSKAALDAFAEVVG
MSMEG_1623|M.smegmatis_MC2_155 VLALLPHWRERRFG-HVVNVSSAGVQAHTPKYSAYLPSKAALDAFSDVVS
Mb3423|M.bovis_AF2122/97 VLALLPHWRERRFG-HVVNVSSAGVQARNPKYSSYLPTKAALDAFADVVA
Rv3391|M.tuberculosis_H37Rv VLALLPHWRERRFG-HVVNVSSAGVQARNPKYSSYLPTKAALDAFADVVA
MAV_4340|M.avium_104 VLALLPHWRERRFG-HVVNVSSAGVQARNPKYSSYLPTKAALDAFADVVG
MMAR_1153|M.marinum_M VLALLPHWRERRFG-HVVNVSSAGVQARNPKYSSYLPSKAALDAFADVVA
MUL_0918|M.ulcerans_Agy99 VLALLPHWRERRFG-HVVNVSSAGVQARNPKYSSYLPSKAALDAFADVVA
MAB_3710|M.abscessus_ATCC_1997 VLALLPHWRERKFG-HVVNVSTAGVQTRNPKYASYIPTKAALDAFADVVA
TH_3707|M.thermoresistible__bu IRGLAPGMRETWG----------VLSEASPLFSVYNASKAALTAVSRVVE
MLBr_00862|M.leprae_Br4923 CRAFGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLR
.: * : * .:*** .: :
Mflv_4896|M.gilvum_PYR-GCK AETLSDHITFTNIHMPLVKTPMIAPSGRLNPAPAISPEHAAAMVVRALVD
Mvan_1527|M.vanbaalenii_PYR-1 TETLSDHITFTNIHMPLVRTPMIAPSGRLNPAPAISAEHAAAMVVRALVD
MSMEG_1623|M.smegmatis_MC2_155 TETLSDHITFTNIHMPLVKTPMIAPSRRLNPVPPISAEHAAAMVVRALVE
Mb3423|M.bovis_AF2122/97 SETLSDHITFTNIHMPLVATPMIVPSRRLNPVRAISAERAAAMVIRGLVE
Rv3391|M.tuberculosis_H37Rv SETLSDHITFTNIHMPLVATPMIVPSRRLNPVRAISAERAAAMVIRGLVE
MAV_4340|M.avium_104 SEVLSDHITFTNIHMPLVRTPMIVPSHRLNPVRAISPERAAAMVVRGLVE
MMAR_1153|M.marinum_M SETLSDHVTFTNIHMPLVATPMIVPSRRLNPTPAISAEHAAAMVVRGLVE
MUL_0918|M.ulcerans_Agy99 SETLSDHVTFTNIHMPLVATPMIVPSRRLNPTPAISAEHAAAMVVRGLVE
MAB_3710|M.abscessus_ATCC_1997 TETVSDHITFTNIHMPLVKTPMITPSHKLNVVRGLTPEHAAAMVVRGLIE
TH_3707|M.thermoresistible__bu TEWSQFGVHSTTLFYPLVKTPMIAPTREYDNLPALSAEEAADWMISAARY
MLBr_00862|M.leprae_Br4923 AELDAVGVGLTTICPGVIDTNIIQSTR-IDTPTAKQERVRDRRGQLDMMF
:* : *.: :: * :* .: : .
Mflv_4896|M.gilvum_PYR-GCK KPARIDTPIGTVADLGMYLTPKLARRVLHQLYLGFPDSAAAQGIAENDRV
Mvan_1527|M.vanbaalenii_PYR-1 KPARIDTPIGTLADVGMYFTPKLARRVLHQLYLGFPDSAAARGITQ-DRS
MSMEG_1623|M.smegmatis_MC2_155 KPPRIDTPLGTFADFGNYLTPRVARRFLHQLYLGYPDSAAARGLAPAEPR
Mb3423|M.bovis_AF2122/97 KPARIDTPLGTLAEAGNYVAPRLSRRILHQLYLGYPDSAAAQGISRPD--
Rv3391|M.tuberculosis_H37Rv KPARIDTPLGTLAEAGNYVAPRLSRRILHQLYLGYPDSAAAQGISRPD--
MAV_4340|M.avium_104 KPARIDTPLGTLAEAGNYFAPRTSRRVLHQLYLGYPDSAAARGVATEPRP
MMAR_1153|M.marinum_M KPARIDTPLGTLAEVGNYVAPKLSRRILHQLYLGYPDSAAAQGISLLD--
MUL_0918|M.ulcerans_Agy99 KPARIDTPLGTLAEIGNYVAPKLSRRILHQLYLGYPDSAAAQGISPLN--
MAB_3710|M.abscessus_ATCC_1997 KPTRIDHPMGTFADIGQYLTPKLSRRVLHQMYLAYPDSKAARGLEPVDDR
TH_3707|M.thermoresistible__bu RPIQIAPRMAVTARALNVVAPGVVNAVMKRQRIQPVFV------------
MLBr_00862|M.leprae_Br4923 RLRRYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTAR
: : . : . . .
Mflv_4896|M.gilvum_PYR-GCK HSATRTP---ERKPWPPRRPVPSVRIP----SAVKQAIRLVPGVHW
Mvan_1527|M.vanbaalenii_PYR-1 R-PRQAP---VRRPKRPSRPMPSVRVP----GPVKRAARLLPGLHW
MSMEG_1623|M.smegmatis_MC2_155 PASTAGPGLPERRPKRPARSRRGVRVPRAVGRPVKRAVRTLPGVHW
Mb3423|M.bovis_AF2122/97 ADRPP-------APRRPRRSARAG-----VPRPLRRLGRLVPGVHW
Rv3391|M.tuberculosis_H37Rv ADRPP-------APRRPRRSARAG-----VPRPLRRLGRLVPGVHW
MAV_4340|M.avium_104 AERIERT-----APRKPRRPVRAVTRGLRTPRPVRRLVRLVPGVHW
MMAR_1153|M.marinum_M -DAQEQR-----AVRRPKRPALAVTG-MRIPRPVKRLVRLTPGVHW
MUL_0918|M.ulcerans_Agy99 -DAQEHR-----PVRRPKRPALAVTG-MRIPRPVKRLVRLAPGVHW
MAB_3710|M.abscessus_ATCC_1997 RDLSTSR----RRPRAAKRVAAVSRLR--VPGPLMKAVRLIPGVHW
TH_3707|M.thermoresistible__bu ----------------------------------------------
MLBr_00862|M.leprae_Br4923 IRVI------------------------------------------