For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGGCSITCLNISEVPNETNRKKNRQAGLDRSIRVIHGSFDDIPEPDSGYDVVWSQDAILHAPDRRKVLEE AFRVLRPGGELIFTDPMQADDVPDGVLQPVYDRLNLRDLGSMRFYA
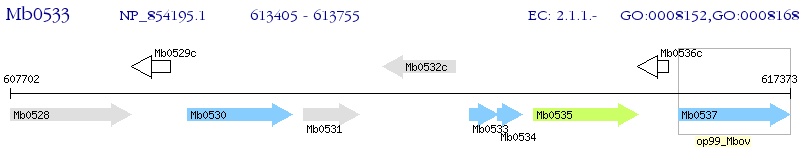
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0533 | - | - | 100% (116) | hypothetical protein Mb0533 |
| M. bovis AF2122 / 97 | Mb1550 | - | 6e-09 | 35.29% (85) | methyltransferase |
| M. bovis AF2122 / 97 | Mb3756 | - | 7e-07 | 27.55% (98) | transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1100 | - | 6e-06 | 26.32% (95) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv0520 | - | 1e-65 | 100.00% (116) | hypothetical protein Rv0520 |
| M. leprae Br4923 | MLBr_01723 | - | 1e-05 | 34.92% (63) | hypothetical protein MLBr_01723 |
| M. abscessus ATCC 19977 | MAB_0018c | - | 6e-07 | 31.25% (80) | putative methyltransferase |
| M. marinum M | MMAR_0861 | - | 2e-51 | 81.42% (113) | glycine-sarcosine methyltransferase |
| M. avium 104 | MAV_3903 | - | 2e-06 | 36.99% (73) | methyltransferase, UbiE/COQ5 family protein |
| M. smegmatis MC2 155 | MSMEG_2350 | - | 1e-07 | 35.80% (81) | hypothetical protein MSMEG_2350 |
| M. thermoresistible (build 8) | TH_2193 | - | 7e-08 | 35.80% (81) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2377 | - | 1e-06 | 28.89% (90) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_2948 | - | 2e-06 | 30.38% (79) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0533|M.bovis_AF2122/97 --------------------------------------------------
Rv0520|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0861|M.marinum_M MTSLSIHQENAELQLNDQIYGDDPLSDRETGLYRAEYVMSFVEKWDELID
MSMEG_2350|M.smegmatis_MC2_155 --------------------------------------------------
TH_2193|M.thermoresistible__bu --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAV_3903|M.avium_104 --------------------------------------------------
Mflv_1100|M.gilvum_PYR-GCK --------------------------------------------------
MUL_2377|M.ulcerans_Agy99 --------------------------------------------------
Mvan_2948|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0018c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0533|M.bovis_AF2122/97 --------------------------------------------------
Rv0520|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0861|M.marinum_M WDARSESEGRFFVDVLRARDKQNILDVATGTGFHSVRLTEAGFNVTSADG
MSMEG_2350|M.smegmatis_MC2_155 --------------------------------------------------
TH_2193|M.thermoresistible__bu --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAV_3903|M.avium_104 --------------------------------------------------
Mflv_1100|M.gilvum_PYR-GCK --------------------------------------------------
MUL_2377|M.ulcerans_Agy99 --------------------------------------------------
Mvan_2948|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0018c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0533|M.bovis_AF2122/97 --------------------------------------------------
Rv0520|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0861|M.marinum_M SAAMLAKAFENGRKRGLILRTVQADWRELNRDIQGKYDAIICLGNSFTHL
MSMEG_2350|M.smegmatis_MC2_155 --------------------------------------------------
TH_2193|M.thermoresistible__bu --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAV_3903|M.avium_104 --------------------------------------------------
Mflv_1100|M.gilvum_PYR-GCK --------------------------------------------------
MUL_2377|M.ulcerans_Agy99 --------------------------------------------------
Mvan_2948|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0018c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0533|M.bovis_AF2122/97 --------------------------------------------------
Rv0520|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0861|M.marinum_M HDEHDRRRALAEFYAALRHDGILILDQRNYDEILDHGFKSKHKYYYCGDQ
MSMEG_2350|M.smegmatis_MC2_155 --------------------------------------------------
TH_2193|M.thermoresistible__bu --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAV_3903|M.avium_104 --------------------------------------------------
Mflv_1100|M.gilvum_PYR-GCK --------------------------------------------------
MUL_2377|M.ulcerans_Agy99 --------------------------------------------------
Mvan_2948|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0018c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0533|M.bovis_AF2122/97 --------------------------------------------------
Rv0520|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0861|M.marinum_M VTAEPEYVDEGLARFKYDFPDGSEYTLNMFPLRKSYVRRLMREAGFTTVR
MSMEG_2350|M.smegmatis_MC2_155 --------------------------------------------------
TH_2193|M.thermoresistible__bu --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAV_3903|M.avium_104 --------------------------------------------------
Mflv_1100|M.gilvum_PYR-GCK --------------------------------------------------
MUL_2377|M.ulcerans_Agy99 --------------------------------------------------
Mvan_2948|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0018c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0533|M.bovis_AF2122/97 --------------------------------------------------
Rv0520|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0861|M.marinum_M TYGDFQESYEETEPDFFIHVAEKSLASEALAEPDIDAALTGTRAEPEAYY
MSMEG_2350|M.smegmatis_MC2_155 -------------------------MGDPDNALPSALPLTGERTIPG---
TH_2193|M.thermoresistible__bu -------------------MSAFSTAGSDADGPISVLPLTGERTVPG---
MLBr_01723|M.leprae_Br4923 ----MTRSTEISANAVPNPHATAEQVAAARKDSKLAQVLYHDWEAENYDE
MAV_3903|M.avium_104 ----MTR----STDAVPTPHATAEQVEAARHDSKLAQVLYHDWEAETYDE
Mflv_1100|M.gilvum_PYR-GCK --------------------------------------------------
MUL_2377|M.ulcerans_Agy99 ----MAFSPAHRLLARLGSTSIYKRVWRYWYPLMTRGLGADKIAFLNWAY
Mvan_2948|M.vanbaalenii_PYR-1 -------------------MAPTNDQAARWNRYWDKKSRTYDR-------
MAB_0018c|M.abscessus_ATCC_199 ------MTTLTSDFTDESTGERLRRWADEWRRMAAVAVSSADGPKTGMVY
Mb0533|M.bovis_AF2122/97 --------------------------------------------------
Rv0520|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0861|M.marinum_M DSDDANAFYSMIWGGEDIHVGCYEDTRDIGAAARKTVDRMVRQLTRFDGD
MSMEG_2350|M.smegmatis_MC2_155 -------------------------------LAEENYWFRRHEVVYQRLA
TH_2193|M.thermoresistible__bu -------------------------------LERENYWFRRHEVVYQRLA
MLBr_01723|M.leprae_Br4923 KWS-----------------------ISYDQRCVDYVRHCFDAIVPDELF
MAV_3903|M.avium_104 KWS-----------------------ISYDQRCIDYARGRFDAIVPEHVL
Mflv_1100|M.gilvum_PYR-GCK -----------------------------------------MLTVDFDRL
MUL_2377|M.ulcerans_Agy99 EEDPP--------------------IDLTLEVSDEPNRDHINMYHRTATH
Mvan_2948|M.vanbaalenii_PYR-1 --------------------------------EIGFFDRHLFGDSRQWVC
MAB_0018c|M.abscessus_ATCC_199 DIVGPQNLFGEESLFINFG-------YWRDHPTTLDEASRDLARLVASSA
Mb0533|M.bovis_AF2122/97 ---------MGGCS----------ITCLNISEVPNETN----------RK
Rv0520|M.tuberculosis_H37Rv ---------MGGCS----------ITCLNISEVPNETN----------RK
MMAR_0861|M.marinum_M TRVLDLGAGYGGCARHLASDYGSSVTCLNISEAQNETN----------RE
MSMEG_2350|M.smegmatis_MC2_155 HRCAGRDVLEAGCGEGYGADLIADVARRVIGLDYDEAT----------VA
TH_2193|M.thermoresistible__bu HRCAGRVVLEAGCGEGYGADLLAEVAAHVIGLDYDAAT----------VA
MLBr_01723|M.leprae_Br4923 TELPYDCALELGCGTGFFLLNLIQSGVARRGSVTDLSPG------MVKIA
MAV_3903|M.avium_104 RELPYDRALELGCGTGFFLLNLIQSGVARRGSVTDLSPG------MVKVA
Mflv_1100|M.gilvum_PYR-GCK GVGAGTKVIDVGCGAGRHTFEAFRRGADVIGFDQSASDLNDVDEILQAMK
MUL_2377|M.ulcerans_Agy99 VELSGKRVLEVSCGHGGGASYLTRTLHPASYTGLDLNRAG--------IK
Mvan_2948|M.vanbaalenii_PYR-1 SQATG-DVLEVAVGTGLNLNFYPDDVALTGIDWSEQMLER---------A
MAB_0018c|M.abscessus_ATCC_199 GFTASDVVVDCGCGYGDQDILWANEFKVKKITGVNIAEEQIAIS----TE
. . .
Mb0533|M.bovis_AF2122/97 KNRQAGLDRSIRVIHGSFDDIPEPDSGYDVVWSQDAILHAPDRRKVLEEA
Rv0520|M.tuberculosis_H37Rv KNRQAGLDRSIRVIHGSFDDIPEPDSGYDVVWSQDAILHAPDRRKVLEEA
MMAR_0861|M.marinum_M RNRNAGLDVKIRVVHGSFEAIPEPDGSYDVVWSQDAILHAADRRKVIEEA
MSMEG_2350|M.smegmatis_MC2_155 HVRAR--YPRVDIRHGNLAELPLPDASVDVVVNFQVIEHLWDQAQFVSEC
TH_2193|M.thermoresistible__bu HVRAR--YPRVDIRHGNLVDLPLADASVDVVVNFQVIEHLWDQEKFIAEC
MLBr_01723|M.leprae_Br4923 TRNGQSLGLDINGRVADAEGIPYDDNTFDLVVGHAVLHHIPDVELSLREV
MAV_3903|M.avium_104 TRNGQSLGLDIDGRVADAEGIPYEDDTFDLVVGHAVLHHIPDVELSLREV
Mflv_1100|M.gilvum_PYR-GCK EQGEAPASAKGEAVKGDALELPYADGTFDCVIASEILEHVPADDKAISEL
MUL_2377|M.ulcerans_Agy99 LCQRRHNLPGLDFVRGDAENLPFEDESFDVVLKVEASHCYPHFSRFLAEV
Mvan_2948|M.vanbaalenii_PYR-1 RRRAAEIGRTATLQQADAHHLPFEDASFDTVVCTFGLCAIPDHAQALTEM
MAB_0018c|M.abscessus_ATCC_199 RVAEAGLSDTISYVKASATDLPFENESCTKVVALESAFHFPSRIDFFREA
.. :* : * . *
Mb0533|M.bovis_AF2122/97 FRVLRPGGELIFT-DPMQADDVP--------------DGVLQPVYDR-LN
Rv0520|M.tuberculosis_H37Rv FRVLRPGGELIFT-DPMQADDVP--------------DGVLQPVYDR-LN
MMAR_0861|M.marinum_M FRVLKAGGELIFT-DPMQADEVP--------------DGVLAPVYDR-LN
MSMEG_2350|M.smegmatis_MC2_155 FRVLRPGGVFLVS-TPNRITFSPG------------RDTPLNPFHTRELN
TH_2193|M.thermoresistible__bu LRVLRPGGQLLVS-TPNRITFTPG------------SDTPVNPFHTRELN
MLBr_01723|M.leprae_Br4923 LRVLKPGGRFVFAGEPTDVGNRYARVFADLTWKVTIRVVQLPGLSAWRRT
MAV_3903|M.avium_104 IRVLRPGGRFVFAGEPTSAGDVYARELSTLTWRIATNVTKLPGLGSWRRP
Mflv_1100|M.gilvum_PYR-GCK VRVLKPGGALAITVPRWLPERIC--------------WALSDEYHANEGG
MUL_2377|M.ulcerans_Agy99 VRVLRPGGYLLYTDLRPSNEIAEW-------------EADLAGSPLRQLS
Mvan_2948|M.vanbaalenii_PYR-1 ARVLRPGGKLVLVDHVQS-------------------TAAPVRAVQRLLE
MAB_0018c|M.abscessus_ATCC_199 LRVLKPGGRMVTADIVPRRTALTA-----------FARSQVARRGWQGAP
***:.** :
Mb0533|M.bovis_AF2122/97 LRDLGSMRFYA---------------------------------------
Rv0520|M.tuberculosis_H37Rv LRDLGSMRFYA---------------------------------------
MMAR_0861|M.marinum_M LADLGSMRFYRETALAVGFEVVDQIDLVHNLGVHYQRVLEELETRRRELE
MSMEG_2350|M.smegmatis_MC2_155 AAELTELLETAGFEVEDTLGVFHGAGLAELDARHGGSIIEAQVQRAVADA
TH_2193|M.thermoresistible__bu AAELTELLTDGGFGLSAMYGVFHGPRLAELDARHGGSIIDAQISRALADE
MLBr_01723|M.leprae_Br4923 QVEIDENSRAAALEWVVDLHTFEPKVLETMATNAGAVQVKTVTEEFTAAM
MAV_3903|M.avium_104 QAELDESSRAAALEAIVDLHTFTPGDLERMAANAGATEVRTVSEEFTAAM
Mflv_1100|M.gilvum_PYR-GCK HIRIYRADELRDKVLAHGLTLTHTHHSHALHSPFWWLKCAVGTEKNDHPA
MUL_2377|M.ulcerans_Agy99 QREINAEVVRGIEKNSHTSRVLVDRNLPAFLRFADRAVGRQLSRYLEGGE
Mvan_2948|M.vanbaalenii_PYR-1 VFTVPLAGEHFLRRPLSHIRSATGLEIDRVERFKLGLVERLVAHKPVAP-
MAB_0018c|M.abscessus_ATCC_199 ANAVPWAVDVQGYRDLLLDMGFARSETWSIANDVYPPLSKFLGKRLRQPD
:
Mb0533|M.bovis_AF2122/97 --------------------------------------------------
Rv0520|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0861|M.marinum_M E-HSSTEYLDKMRVG----------LRNWVDSAR------------EGHL
MSMEG_2350|M.smegmatis_MC2_155 P-WDEQLLADVAAVR----------TDDFDLTPAAERD-------IDDSL
TH_2193|M.thermoresistible__bu P-WPAELLADVASVR----------TEDFDLIRCDEDDRDRDGHNIDDSL
MLBr_01723|M.leprae_Br4923 LGWPLRTFESTMPPSKLGWGWARFAFTSWKTLSWVDANVWRRVIPKSWFY
MAV_3903|M.avium_104 FGWPVRTFEASVPPGRLGWGWAKFAFNGWKTLSWVDANIWRRVVPKGWFY
Mflv_1100|M.gilvum_PYR-GCK VAAYHKLLVWDMMSQP------------WLTRTAEAALNPLIGKSVALYF
MUL_2377|M.ulcerans_Agy99 LSYRMYCFTKDFAASR----------------------------------
Mvan_2948|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0018c|M.abscessus_ATCC_199 MKHVNPLLRAQFNSVG--------------------------MRLFSLHS
Mb0533|M.bovis_AF2122/97 -----------
Rv0520|M.tuberculosis_H37Rv -----------
MMAR_0861|M.marinum_M AWGIQHFRKPD
MSMEG_2350|M.smegmatis_MC2_155 DLVAIAVRP--
TH_2193|M.thermoresistible__bu DLVAIAVRP--
MLBr_01723|M.leprae_Br4923 NIIITGVKPS-
MAV_3903|M.avium_104 NVMVTGVKPT-
Mflv_1100|M.gilvum_PYR-GCK RKPADTDA---
MUL_2377|M.ulcerans_Agy99 -----------
Mvan_2948|M.vanbaalenii_PYR-1 -----------
MAB_0018c|M.abscessus_ATCC_199 DYIVAVAHKA-