For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTELRPFYEESQSIYDVSDEFFSLFLDPTMAYTCAYFEREDMTLEEAQNAKFDLALDKLHLEPGMTLLDI GCGWGGGLQRAIENYDVNVIGITLSRNQFEYSKAKLAKIPTERSVQVRLQGWDEFTDKVDRIVSIGAFEA FKMERYAAFFERSYDILPDDGRMLLHTILTYTQKQMHEMGVKVTMSDVRFMKFIGEEIFPGGQLPAQEDI FKFAQAADFSVEKVQLLQQHYARTLNIWAANLEANKDRAIALQSEEIYNKYMHYLTGCEHFFRKGISNVG QFTLTK
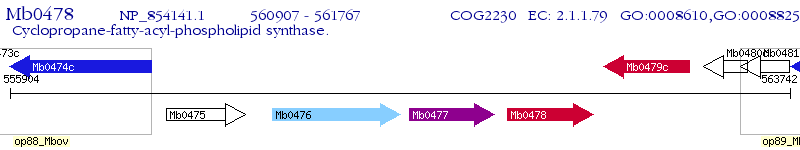
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0478 | umaA1 | - | 100% (286) | mycolic acid synthase |
| M. bovis AF2122 / 97 | Mb0664c | mmaA1 | e-120 | 69.58% (286) | methoxy mycolic acid synthase |
| M. bovis AF2122 / 97 | Mb3424c | cmaA1 | 4e-91 | 56.69% (284) | cyclopropane-fatty-acyl-phospholipid synthase 1 CMAA1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3304 | - | 4e-96 | 59.15% (284) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. tuberculosis H37Rv | Rv0469 | umaA | 1e-167 | 100.00% (286) | mycolic acid synthase |
| M. leprae Br4923 | MLBr_01900 | mmaA1 | 1e-117 | 67.83% (286) | methyl mycolic acid synthase 1 |
| M. abscessus ATCC 19977 | MAB_4088c | - | 1e-116 | 67.82% (289) | mycolic acid synthase UmaA1 |
| M. marinum M | MMAR_0794 | umaA | 1e-152 | 89.86% (286) | mycolic acid synthase UmaA |
| M. avium 104 | MAV_4680 | - | 1e-143 | 82.87% (286) | methoxy mycolic acid synthase 1 |
| M. smegmatis MC2 155 | MSMEG_0913 | - | 1e-122 | 71.58% (285) | methoxy mycolic acid synthase 1 |
| M. thermoresistible (build 8) | TH_3763 | - | 1e-121 | 71.83% (284) | PUTATIVE Cyclopropane-fatty-acyl-phospholipid synthase |
| M. ulcerans Agy99 | MUL_4538 | umaA | 1e-152 | 89.51% (286) | mycolic acid synthase UmaA |
| M. vanbaalenii PYR-1 | Mvan_0803 | - | 1e-120 | 71.48% (284) | cyclopropane-fatty-acyl-phospholipid synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0478|M.bovis_AF2122/97 -------MTELRPFYEESQSIYDVSDEFFSLFLD-PTMAYTCAYFEREDM
Rv0469|M.tuberculosis_H37Rv -------MTELRPFYEESQSIYDVSDEFFSLFLD-PTMAYTCAYFEREDM
MMAR_0794|M.marinum_M -------MTELRPFYEESQSIYDVSDEFFALFLD-PTMAYTCAYFERDDM
MUL_4538|M.ulcerans_Agy99 -------MTELRPFYEESQSIYDVSDEFFALFLD-PTMAYTCAYFERDDM
MAV_4680|M.avium_104 ----MPRMTGLEPYYEESQSIYDVSDEFFALFLD-PTMAYTCAFFERDDM
MSMEG_0913|M.smegmatis_MC2_155 ---MSNVESELAPYYEESQSIYDISNEFFALFLG-PTMGYTCGYYEREDM
Mvan_0803|M.vanbaalenii_PYR-1 ---MSNAVPDLSPYYEESQSIYDVSNEFFALFLG-PTMGYTCSYFEREDM
MAB_4088c|M.abscessus_ATCC_199 ---MS----DLKPYYEESQSIYDISDEFYGLFLDEETMGYTCAYFERDDL
TH_3763|M.thermoresistible__bu ---MTNTSAELEPFFEESQSIYDVSDDFFALFLG-PTMGYTCAYFERDDM
MLBr_01900|M.leprae_Br4923 -------MAKLRPYYEESQSAYDISDDFFALFLD-PTWVYTCAYFERDDM
Mflv_3304|M.gilvum_PYR-GCK MSDTPNGSADLQPHFDDVQSHYDLSDDFYRLFLD-PTQTYSCAYFERDDM
* *.::: ** **:*::*: ***. * *:*.::**:*:
Mb0478|M.bovis_AF2122/97 TLEEAQNAKFDLALDKLHLEPGMTLLDIGCGWGGGLQRAIENYDVNVIGI
Rv0469|M.tuberculosis_H37Rv TLEEAQNAKFDLALDKLHLEPGMTLLDIGCGWGGGLQRAIENYDVNVIGI
MMAR_0794|M.marinum_M TLEEASNAKFDLALGKLNLEPGMTLLDIGCGWGGALQRAVEKYDVNVIGI
MUL_4538|M.ulcerans_Agy99 TLEEASNAKFDLALGKLNLEPGMTLLDIGCGWGGALQRAVEKYDVNVIGI
MAV_4680|M.avium_104 TLEEAQTAKFDLALGKLNLEPGMTLLDIGCGWGGALKRALEKYDVNVIGI
MSMEG_0913|M.smegmatis_MC2_155 TLDESQNAKFDLALGKLDLKPGMTLLDVGCGWGGALERAVTKYDVNVIGI
Mvan_0803|M.vanbaalenii_PYR-1 TLDEAQIAKFDLALGKLNLEPGMTLLDVGCGWGGALRRAVEKYDVNVIGI
MAB_4088c|M.abscessus_ATCC_199 TLAEAQIAKFDLALGKLNLEPGMTVLDIGCGWGACLDRAMRKFDVNVIGI
TH_3763|M.thermoresistible__bu TLDEAQIAKFDLALGKLNLEPGMTLLDIGCGWGGALQLALEKYDVNVIGI
MLBr_01900|M.leprae_Br4923 TLEEAQLAKLDLALDKLNLAPGMTLLDVGCGWGGALVRAVEKYDVNVIGL
Mflv_3304|M.gilvum_PYR-GCK TLEEAQRAKIDLALGKLGLEPGMTLLDIGCGWGGTLLRALEKYDVNVIGL
** *:. **:****.** * ****:**:*****. * *: ::******:
Mb0478|M.bovis_AF2122/97 TLSRNQFEYSKAKLAKIP--TERSVQVRLQGWDEFTDKVDRIVSIGAFEA
Rv0469|M.tuberculosis_H37Rv TLSRNQFEYSKAKLAKIP--TERSVQVRLQGWDEFTDKVDRIVSIGAFEA
MMAR_0794|M.marinum_M TLSRNQFEYSKAKMAKIP--TERTVEVRLQGWEEFEDKVDRIVTIGAFEA
MUL_4538|M.ulcerans_Agy99 TLSRNQFDYSKAKMAKIP--TERTVEVRLQGWEEFEDKVDRIVTIGAFEA
MAV_4680|M.avium_104 TLSRNQFEYSKKLLAGFP--TDRNIEVRLQGWEEFDDKVDRIVTIGAFEA
MSMEG_0913|M.smegmatis_MC2_155 TLSKAQSDYARERLAKI--ETNRSVEVRLQGWEEFNEPVDRIVSIGAFEA
Mvan_0803|M.vanbaalenii_PYR-1 TLSRKQFEYSKALMAGL--DTERKAEVRLQGWEEFDEPVDRIVSIGAFEA
MAB_4088c|M.abscessus_ATCC_199 TLSKNQSEYSRKRLAQVAAETGRSAEIRMQGWEEFNDKVDRIVTIGAFEA
TH_3763|M.thermoresistible__bu TLSRNQYEYSKARLAQI--DTARTAEVRLQGWEQFDEPVDRIVSIGAFEA
MLBr_01900|M.leprae_Br4923 TLSRNHCARSKARLAEIL--TKRHAEARLQGWEEFEEKVDRIVSFEALDA
Mflv_3304|M.gilvum_PYR-GCK TLSRNQQAHVQKVLDEHD--SPRSKTVLLQGWEQFDQPVDRIVSIGAFEH
***: : : : : * :***::* : *****:: *::
Mb0478|M.bovis_AF2122/97 FKMERYAAFFERSYDILPDDGRMLLHTILTYTQKQMHEMGVKVTMSDVRF
Rv0469|M.tuberculosis_H37Rv FKMERYAAFFERSYDILPDDGRMLLHTILTYTQKQMHEMGVKVTMSDVRF
MMAR_0794|M.marinum_M FKMERYAAFFERSYDILPDDGRMLLHTILTYTQKQFHEMGVKVTMSDVRF
MUL_4538|M.ulcerans_Agy99 FKMERYAAFFERSYDILPDDGRMLLHTILTYTQKQFHEMGVKVTMSDVRF
MAV_4680|M.avium_104 FKMERYSAFFDRAYDILPDDGRMLLHTILTYTQKQQHERGVSITMSDLRF
MSMEG_0913|M.smegmatis_MC2_155 FKAERYPVFFERAYSILPDDGRMLLHTILAHTQKFFRENGIKLTISDLKF
Mvan_0803|M.vanbaalenii_PYR-1 FKAERYPLFFERAYDILPADGRMLLHTILAHTQKFFRDNGIKLTISDLKF
MAB_4088c|M.abscessus_ATCC_199 FKQERYPIFFERAYDILPNDGRMLLHTILAHTQQFFRENGIAITISDLKF
TH_3763|M.thermoresistible__bu FKKPRYAAFFEMAHKALPADGRMLLHTILAHPQTYFREAGIKITISDLKF
MLBr_01900|M.leprae_Br4923 FKKERYPAFFERSYNILPDDGRMLLHSLFTYDRRWLHEQGIPLTMGDLRF
Mflv_3304|M.gilvum_PYR-GCK FGSDRYDEFFRRAYSVLPDDGVMLLHTIVKPSDEEFAERKLPLTMTKLKF
* ** ** ::. ** ** ****::. : : :*: .::*
Mb0478|M.bovis_AF2122/97 MKFIGEEIFPGGQLPAQEDIFKFAQAADFSVEKVQLLQQHYARTLNIWAA
Rv0469|M.tuberculosis_H37Rv MKFIGEEIFPGGQLPAQEDIFKFAQAADFSVEKVQLLQQHYARTLNIWAA
MMAR_0794|M.marinum_M MRFIGQEIFPGGQLPAQEDIFNFAQAAGFSVERAQLLQQHYARTLNIWAA
MUL_4538|M.ulcerans_Agy99 MRFIGQEIFPGGQLPAQEDIFNFAQAAGFSVERAQLLQQHYARTLNIWAA
MAV_4680|M.avium_104 MRFIGQEIFPGGQLPAQEDIFKFGEAAGFSVERVQLLREHYARTLNIWAA
MSMEG_0913|M.smegmatis_MC2_155 MKFIGEEIFPGGQLPAVEDIEKLAADSGFKLERIHLLQPHYARTLDMWAA
Mvan_0803|M.vanbaalenii_PYR-1 MRFIGTEIFPGGQLPAVEDIEKLAADSGFTLERAHLLRPHYARTLDMWAA
MAB_4088c|M.abscessus_ATCC_199 MKFIGEEIFPGGQLPAVEDIEKLAADSGFNLERVHLLQPHYAKTLDIWAQ
TH_3763|M.thermoresistible__bu MQFIGREIFPGGELPAEGDVVEFATGAGFSVERIQLLRPHYARTLDIWAE
MLBr_01900|M.leprae_Br4923 LKFLRESIFPGGELPSQPDIVDNAEAAGFSVEQIQLMQPHYARTLDMWAT
Mflv_3304|M.gilvum_PYR-GCK FKFIMDEIFPGGMLPKVDDVEKHATKASFKVNRIQPLRLHYARTLQIWAA
::*: .***** ** *: . . :.*.::: : :: ***:**::**
Mb0478|M.bovis_AF2122/97 NLEANKDRAIALQSEEIYNKYMHYLTGCEHFFRKGISNVGQFTLTK-
Rv0469|M.tuberculosis_H37Rv NLEANKDRAIALQSEEIYNKYMHYLTGCEHFFRKGISNVGQFTLTK-
MMAR_0794|M.marinum_M NLEARKEEAIAMQSQEIYDKYMHYLTGCENFFRKGISNVGQFTLVK-
MUL_4538|M.ulcerans_Agy99 NLEARKEEAIAMQSQEIYDKYMHYLTGCENFFRKGISNVGQFTLVK-
MAV_4680|M.avium_104 NLEANKDKAIAIQSQAVYDRYMHYLTGCENFFRKGISNVGQFTLVK-
MSMEG_0913|M.smegmatis_MC2_155 NLEAKKDEAIAMQGQEVYDRYMKYLTGCADFFRRGITNVGQFTLVK-
Mvan_0803|M.vanbaalenii_PYR-1 NLEANRQEATRITSPEVYERYMHYLTGCADFFRRGITNVGQFTLVKG
MAB_4088c|M.abscessus_ATCC_199 NLEQRREEAIAIQSQEVYDRFMKYLTGCADFFRRGVTNVGQFTLVK-
TH_3763|M.thermoresistible__bu NLAANREPAIAMQSEEVYDRYYRYLTGCADFFRRGITNVGQFTLTKG
MLBr_01900|M.leprae_Br4923 NLAAARDHALAIQPEEIYDNFMHYLTGCADRFRRGLINVAQFTLTK-
Mflv_3304|M.gilvum_PYR-GCK SLESRRDDAIAIQSEEVYDRYMKYLTGCAELFTEGYTDVCQFTLAK-
.* :: * : :*:.: :***** . * .* :* ****.*