For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGDLSIQNLVVEYYSGGYALRPINGLNLDVAAGSLVMLLGPSGCGKTTLLSCLGGILRPKSGAIKFDEVD ITTLQGAELANYRRNKVGIVFQAFNLVPSLTAVENVMVPLRSAGMSRRASRRRAEELLARVNLAERMNHR PGDLSGGQQQRVAVARAIALDPPLILADEPTAHLDFIQVEEVLRLIRELADGERVVVVATHDSRMLPMAD RVVELTPDFAETNRPPETVHLQAGEVLFEQSTMGDLIYVVSEGEFEIVHDWPTAVRNWSRLPGRGITSAR
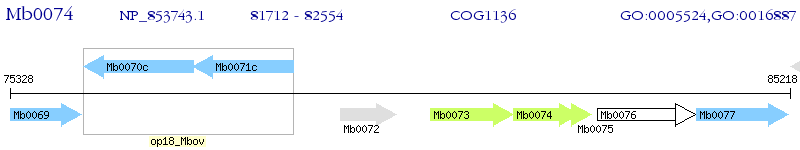
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0074 | - | - | 100% (280) | glutamine-transport ATP-binding protein ABC transporter GlnQ |
| M. bovis AF2122 / 97 | Mb2593 | glnQ | e-119 | 82.31% (260) | glutamine-transport ATP-binding protein ABC transporter GlnQ |
| M. bovis AF2122 / 97 | Mb1012 | - | 2e-38 | 39.44% (213) | adhesion component transport ATP-binding protein ABC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4127 | - | 2e-42 | 44.28% (201) | ABC transporter related |
| M. tuberculosis H37Rv | Rv0073 | - | 1e-144 | 94.91% (275) | glutamine-transport ATP-binding protein ABC transporter |
| M. leprae Br4923 | MLBr_00669 | ftsE | 6e-29 | 35.90% (195) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_4671c | - | 3e-42 | 43.06% (216) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_0216 | - | 1e-131 | 85.82% (275) | glutamine-transport ATP-binding protein ABC transporter |
| M. avium 104 | MAV_4425 | - | 1e-45 | 45.58% (215) | ABC-type transporter, ATPase component |
| M. smegmatis MC2 155 | MSMEG_2524 | - | 2e-41 | 43.28% (201) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_2538 | - | 6e-37 | 40.65% (214) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_0674 | - | 1e-39 | 40.65% (214) | ABC transporter, ATP-binding protein |
| M. vanbaalenii PYR-1 | Mvan_1004 | - | 3e-46 | 44.86% (214) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0074|M.bovis_AF2122/97 ------------------MGDLSIQNLVVEYYSGGYALRPINGLNLDVAA
Rv0073|M.tuberculosis_H37Rv ------------------MGDLSIQNLVVEYYSGGYALRPINGLNLDVAA
MMAR_0216|M.marinum_M ------------------MGDLSIQDLVVEYYSGGYALRPINGLNLDVEA
MAV_4425|M.avium_104 -------------------MTIEMAGVVREYRVGGQPVRALDRISLQLAG
Mvan_1004|M.vanbaalenii_PYR-1 --------------------MLEMTDVTKTYRVGGQTVRALDGITLSLCG
MUL_0674|M.ulcerans_Agy99 --------MTRAQAPTSPRGGVSLHGVDKTFGTAAASVHALRRIDLDIEP
Mflv_4127|M.gilvum_PYR-GCK MTVVDETPDTPTGPVADPAPVIEISDVWKLHKLGDEVVKALMAAELTVMP
MSMEG_2524|M.smegmatis_MC2_155 MTALDDT-EAPADNEDSRKPVIEISDVWKLHKLGDEVVKALKAAELQVMP
MAB_4671c|M.abscessus_ATCC_199 -------MNSPERPNVSPEPVLQLKGVAHQYGTGAAAVHALRGLDLTVNT
MLBr_00669|M.leprae_Br4923 --------------------MITLDHVSKKYKS--LARPALDNVNVKIDK
TH_2538|M.thermoresistible__bu --------------------MIELTDLTKVYGCGEHRTVVLDRINFRVET
: : : . : . :
Mb0074|M.bovis_AF2122/97 GSLVMLLGPSGCGKTTLLSCLGGILRPKSGAIKFDEVDITTLQGAELANY
Rv0073|M.tuberculosis_H37Rv GSLVMLLGPSGCGKTTLLSCLGGILRPKSGAIKFDEVDITTLQGAELANY
MMAR_0216|M.marinum_M GSLVILLGPSGCGKTTLLSCLGGILRPKSGAIKFEDIDITTLEGTALAKY
MAV_4425|M.avium_104 GQFVAVVGPSGAGKSTLLHLLGALDSPDSGSILFDGEEIGRLGDEAQSRF
Mvan_1004|M.vanbaalenii_PYR-1 GEFVSVVGPSGAGKSTLLHLLGALDRPDAGSIRFQGAEIAALDDEAQSEF
MUL_0674|M.ulcerans_Agy99 GELVAIVGESGSGKTTLLNVIGGIEEANSGSINVAGQDISGCRPRALSKF
Mflv_4127|M.gilvum_PYR-GCK GEFVCLMGPSGSGKSTLLNIIGGLDRPTKGNVRIAGTDTATLTESQYAAL
MSMEG_2524|M.smegmatis_MC2_155 GEFVCLMGPSGSGKSTLLNIIGGLDRPTKGTVKISGQDTAQLTESQFAAL
MAB_4671c|M.abscessus_ATCC_199 GEVVLVVGPSGGGKTTALLVMGLLLTPVSGLVRIGGRDVGELAERDRAHI
MLBr_00669|M.leprae_Br4923 GEFVFLIGPSGSGKSTFMRLLLGAETPTSGDVRVSKFHVNKLPGRHIPRL
TH_2538|M.thermoresistible__bu GEVLAVVGPSGAGKSTLAQCINLLTRPTSGSVVVNGEDLTTLSTHRLRVA
*..: ::* ** **:* : . * : . .
Mb0074|M.bovis_AF2122/97 RRNKVGIVFQAFNLVPSLTAVENVMVPLRSAGMSRRASRRRAEELLARVN
Rv0073|M.tuberculosis_H37Rv RRNKVGIVFQAFNLVPSLTAVENVMVPLRSAGMSRRASRRRAEELLARVN
MMAR_0216|M.marinum_M RRDKVGIVFQAFNLVPSLTALENVMVPLRSAGMSRAAARARAEELLKRVN
MAV_4425|M.avium_104 RHRRVGFVFQFFNLLPTLSAWENVAIPRLLDGVRLGRARPDALRLLERVG
Mvan_1004|M.vanbaalenii_PYR-1 RRHSVGFVFQFFNLLPTMTAWENVAVPKLLDGSRMAKVKSRALDLLAVVG
MUL_0674|M.ulcerans_Agy99 RRDHVGFIFQFFNLIPTLTAKENVEVIVELTG---RGDRRRATELLDAVE
Mflv_4127|M.gilvum_PYR-GCK RHDTIGFIFQSYNLIPFLSAVENVELPLMFEPYDRKALRKRAIELLELVG
MSMEG_2524|M.smegmatis_MC2_155 RHDTIGFIFQSYNLIPFLSAVENVELPLMFEPYDRKSLRTRATELLEMVG
MAB_4671c|M.abscessus_ATCC_199 RLEKLGFLFQEYNLLNALTSAENVALPLRYAGVSKDKAMSRARGLLADVG
MLBr_00669|M.leprae_Br4923 R-QVIGCVFQDFRLLQQKTVYENVAFALEVIGRRSDAINQVVPDVLETVG
TH_2538|M.thermoresistible__bu R-RRIGTVFQSSGLLERRTAAENVALPLEYLGVTAAETRKRVAELLDRVG
* :* :** *: : *** . . :* *
Mb0074|M.bovis_AF2122/97 LAERMNHRPGDLSGGQQQRVAVARAIALDPPLILADEPTAHLDFIQVEEV
Rv0073|M.tuberculosis_H37Rv LAERMNHRPGDLSGGQQQRVAVARAIALDPPLILADEPTAHLDFIQVEEV
MMAR_0216|M.marinum_M LEERMKHRPGDLSGGQQQRVAVARAIALDPPLILADEPTAHLDFIQVEEV
MAV_4425|M.avium_104 LGNRTEHRPAELSGGQMQRVAVARALMMDPPLILADEPTGNLDSSTGAAI
Mvan_1004|M.vanbaalenii_PYR-1 LGDRAEHRPSELSGGQMQRVAVARALMMDPPLILADEPTGNLDTKTGAAI
MUL_0674|M.ulcerans_Agy99 LADRAGSFPGQLSGGQQQRVAIARALITNPDLLLADEPMGALDVATGRRV
Mflv_4127|M.gilvum_PYR-GCK LGHRINHQPTRMSGGEQQRTAIARSLISNPALVLADEPTANLDHRTGETV
MSMEG_2524|M.smegmatis_MC2_155 LGHRIHHQPTKMSGGEQQRTAIARSLISNPTLVLADEPTANLDHRTGETV
MAB_4671c|M.abscessus_ATCC_199 MEHRTDHRPPALSGGEKQRVAAARALVARPGLVLADEPTANLDSATGKRV
MLBr_00669|M.leprae_Br4923 LSGKANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDI
TH_2538|M.thermoresistible__bu LAARANHYPFQLSGGQRQRVGIARALALRPSVLLSDEATAGLDPTTTASV
: : * :***: **.. **:: * ::*:**. . ** :
Mb0074|M.bovis_AF2122/97 LRLIRELADGE---RVVVVATHDSRMLP-MADRVVELTPDFAETNRPPET
Rv0073|M.tuberculosis_H37Rv LRLIRELADGE---RVVVVATHDSRMLP-MADRVVELTPDFAETNRPPET
MMAR_0216|M.marinum_M LRLIRELADGD---RVVVVATHDSRMLP-MADRVVELTPDFAETNRPPEV
MAV_4425|M.avium_104 LELLAEVAHEGGGDRLVVMVTHNSDAAA-ATDRVITLQ------------
Mvan_1004|M.vanbaalenii_PYR-1 MALLTDIAHQD--ERSVVMVTHNLGAAS-ATDRVITLT------------
MUL_0674|M.ulcerans_Agy99 LELLQQTNRSG---LGVIMVTHNRAAAD-VADPIIQMR------------
Mflv_4127|M.gilvum_PYR-GCK VRMLRDLCSTM--GVTVVASTHDPTVAD-EASRVVRMK------------
MSMEG_2524|M.smegmatis_MC2_155 VRMLRDLCLTL--GVTVVASTHDPTVAD-EASRVVRMR------------
MAB_4671c|M.abscessus_ATCC_199 AAQLADAARSQ--GAAVVIVTHDLRLTD-IADRVLHLE------------
MLBr_00669|M.leprae_Br4923 MDLLERINRTG---TTVLMATHDHHIVDSMRQRVVELS------------
TH_2538|M.thermoresistible__bu VELLRELRDDLN--LSIVFITHEMDTVLKIADSVARLD------------
: :: **: . : :
Mb0074|M.bovis_AF2122/97 VHLQAGEVLFEQSTMGDLIYVVSEGEFEIVHDWPTAVRNWSRLPGRG---
Rv0073|M.tuberculosis_H37Rv VHLQAGEVLFEQSTMGDLIYVVSEGEFEIVHELADGGEELVKVAGPGDYF
MMAR_0216|M.marinum_M VELKSGDVLFEQSTMGDLIYVVSAGEFEIVHELADGGEELVKVAGPGEYF
MAV_4425|M.avium_104 -----------DGRVGSDVLAVAG--------------------------
Mvan_1004|M.vanbaalenii_PYR-1 -----------DGRIGSDVLAGGELT------------------------
MUL_0674|M.ulcerans_Agy99 -----------DGAIVEQRRNSAPAEASSVRW------------------
Mflv_4127|M.gilvum_PYR-GCK -----------DGQIVN---------------------------------
MSMEG_2524|M.smegmatis_MC2_155 -----------DGQIIN---------------------------------
MAB_4671c|M.abscessus_ATCC_199 -----------DGVLLEKENQ-----------------------------
MLBr_00669|M.leprae_Br4923 -----------LGRLVRD--------------------------------
TH_2538|M.thermoresistible__bu -----------HGRIVEAGRLVDLLTDPASALGADLRPQGAAAPPAPPXX
. :
Mb0074|M.bovis_AF2122/97 -------------ITSAR--------------------------------
Rv0073|M.tuberculosis_H37Rv GEIGVLFHLPRSATVRARSDATAVGYTVQAFRERLGVGGLRDLIEHRALA
MMAR_0216|M.marinum_M GEMGVLFHMPRSATVRARTDATAVGYTAQAFRERLGIGGMRDLIEHRELS
MAV_4425|M.avium_104 --------------------------------------------------
Mvan_1004|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_0674|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4127|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2524|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4671c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00669|M.leprae_Br4923 -EWCGIYGMDR---------------------------------------
TH_2538|M.thermoresistible__bu XRLRGVAGPDHPHRTGAVRAAGRTGPAAARRCGGGTAVDAGSDRRVADRA
Mb0074|M.bovis_AF2122/97 ---------------------
Rv0073|M.tuberculosis_H37Rv ND-------------------
MMAR_0216|M.marinum_M ND-------------------
MAV_4425|M.avium_104 ---------------------
Mvan_1004|M.vanbaalenii_PYR-1 ---------------------
MUL_0674|M.ulcerans_Agy99 ---------------------
Mflv_4127|M.gilvum_PYR-GCK ---------------------
MSMEG_2524|M.smegmatis_MC2_155 ---------------------
MAB_4671c|M.abscessus_ATCC_199 ---------------------
MLBr_00669|M.leprae_Br4923 ---------------------
TH_2538|M.thermoresistible__bu GRRPGRRDDAVAAPRRRPIRP