For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLEMTDVTKTYRVGGQTVRALDGITLSLCGGEFVSVVGPSGAGKSTLLHLLGALDRPDAGSIRFQGAEIA ALDDEAQSEFRRHSVGFVFQFFNLLPTMTAWENVAVPKLLDGSRMAKVKSRALDLLAVVGLGDRAEHRPS ELSGGQMQRVAVARALMMDPPLILADEPTGNLDTKTGAAIMALLTDIAHQDERSVVMVTHNLGAASATDR VITLTDGRIGSDVLAGGELT
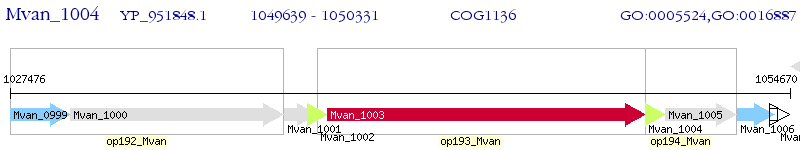
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1004 | - | - | 100% (230) | ABC transporter-related protein |
| M. vanbaalenii PYR-1 | Mvan_2216 | - | 7e-50 | 41.10% (219) | ABC transporter-related protein |
| M. vanbaalenii PYR-1 | Mvan_3884 | - | 3e-36 | 40.00% (220) | ABC transporter-related protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2593 | glnQ | 3e-48 | 45.41% (218) | glutamine-transport ATP-binding protein ABC transporter GlnQ |
| M. gilvum PYR-GCK | Mflv_4127 | - | 2e-50 | 41.55% (219) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2564 | glnQ | 3e-48 | 45.41% (218) | glutamine-transport ATP-binding protein ABC transporter GlnQ |
| M. leprae Br4923 | MLBr_00669 | ftsE | 1e-33 | 38.77% (227) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_4671c | - | 8e-46 | 45.16% (217) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_0922 | - | 1e-48 | 48.83% (213) | ABC transporter, ATP-binding protein |
| M. avium 104 | MAV_4425 | - | 9e-90 | 73.01% (226) | ABC-type transporter, ATPase component |
| M. smegmatis MC2 155 | MSMEG_2524 | - | 2e-49 | 41.10% (219) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4101 | - | 5e-44 | 42.34% (222) | PUTATIVE putative ABC transporter, ATP-binding protein |
| M. ulcerans Agy99 | MUL_0674 | - | 1e-45 | 46.95% (213) | ABC transporter, ATP-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1004|M.vanbaalenii_PYR-1 --------------------MLEMTDVTKTYRVGGQTVRALDGITLSLCG
MAV_4425|M.avium_104 -------------------MTIEMAGVVREYRVGGQPVRALDRISLQLAG
Mb2593|M.bovis_AF2122/97 ------------------MGGLTISDLVVEYSSGGYAVRPIDGLSLDVAP
Rv2564|M.tuberculosis_H37Rv ------------------MGGLTISDLVVEYSSGGYAVRPIDGLSLDVAP
Mflv_4127|M.gilvum_PYR-GCK MTVVDETPDTPTGPVADPAPVIEISDVWKLHKLGDEVVKALMAAELTVMP
MSMEG_2524|M.smegmatis_MC2_155 MTALDDT-EAPADNEDSRKPVIEISDVWKLHKLGDEVVKALKAAELQVMP
MAB_4671c|M.abscessus_ATCC_199 -------MNSPERPNVSPEPVLQLKGVAHQYGTGAAAVHALRGLDLTVNT
MMAR_0922|M.marinum_M --------MTRAQAPTSPRGGVSLHGVDKTFGTAAASVHALRGIDLDIEP
MUL_0674|M.ulcerans_Agy99 --------MTRAQAPTSPRGGVSLHGVDKTFGTAAASVHALRRIDLDIEP
MLBr_00669|M.leprae_Br4923 --------------------MITLDHVSKKYKS--LARPALDNVNVKIDK
TH_4101|M.thermoresistible__bu ---------VTDSGEDRDTAVIEARDLCKSFGQGAAETPILKHVDVRISP
: : . : : :
Mvan_1004|M.vanbaalenii_PYR-1 GEFVSVVGPSGAGKSTLLHLLGALDRPDAGSIRFQGAEIAALDDEAQSEF
MAV_4425|M.avium_104 GQFVAVVGPSGAGKSTLLHLLGALDSPDSGSILFDGEEIGRLGDEAQSRF
Mb2593|M.bovis_AF2122/97 GSLVILLGPSGCGKTTLLSCLGGILRPKSGSIKFDDVDITTLEGAALAKY
Rv2564|M.tuberculosis_H37Rv GSLVILLGPSGCGKTTLLSCLGGILRPKSGSIKFDDVDITTLEGAALAKY
Mflv_4127|M.gilvum_PYR-GCK GEFVCLMGPSGSGKSTLLNIIGGLDRPTKGNVRIAGTDTATLTESQYAAL
MSMEG_2524|M.smegmatis_MC2_155 GEFVCLMGPSGSGKSTLLNIIGGLDRPTKGTVKISGQDTAQLTESQFAAL
MAB_4671c|M.abscessus_ATCC_199 GEVVLVVGPSGGGKTTALLVMGLLLTPVSGLVRIGGRDVGELAERDRAHI
MMAR_0922|M.marinum_M GELVAIVGESGSGKTTLLNVIGGIEEANSGSINVAGQDISGCRPRALSKF
MUL_0674|M.ulcerans_Agy99 GELVAIVGESGSGKTTLLNVIGGIEEANSGSINVAGQDISGCRPRALSKF
MLBr_00669|M.leprae_Br4923 GEFVFLIGPSGSGKSTFMRLLLGAETPTSGDVRVSKFHVNKLPGRHIPRL
TH_4101|M.thermoresistible__bu GEFVVLIGPSGSGKSTLLSILGLLEPPSSGRLFVDQQDVSGLSRRQLARL
*..* ::* ** **:* : : . * : . . .
Mvan_1004|M.vanbaalenii_PYR-1 RRHSVGFVFQFFNLLPTMTAWENVAVPKLLDGSRMAKVKSRALDLLAVVG
MAV_4425|M.avium_104 RHRRVGFVFQFFNLLPTLSAWENVAIPRLLDGVRLGRARPDALRLLERVG
Mb2593|M.bovis_AF2122/97 RRDKVGIVFQAFNLVSSLTALENVMVPLRAAGVSRAAARKRAEDLLIRVN
Rv2564|M.tuberculosis_H37Rv RRDKVGIVFQAFNLVSSLTALENVMVPLRAAGVSRAAARKRAEDLLIRVN
Mflv_4127|M.gilvum_PYR-GCK RHDTIGFIFQSYNLIPFLSAVENVELPLMFEPYDRKALRKRAIELLELVG
MSMEG_2524|M.smegmatis_MC2_155 RHDTIGFIFQSYNLIPFLSAVENVELPLMFEPYDRKSLRTRATELLEMVG
MAB_4671c|M.abscessus_ATCC_199 RLEKLGFLFQEYNLLNALTSAENVALPLRYAGVSKDKAMSRARGLLADVG
MMAR_0922|M.marinum_M RRDHVGFIFQFFNLIPTLTAKENVEVIVELTG---RGDRRRATELLDAVG
MUL_0674|M.ulcerans_Agy99 RRDHVGFIFQFFNLIPTLTAKENVEVIVELTG---RGDRRRATELLDAVE
MLBr_00669|M.leprae_Br4923 R-QVIGCVFQDFRLLQQKTVYENVAFALEVIGRRSDAINQVVPDVLETVG
TH_4101|M.thermoresistible__bu RGRKLGYVFQSFNLLAGLSVAENVMLSGLLVGESGRAQYERAVELLDEFG
* :* :** :.*: : *** . . :* .
Mvan_1004|M.vanbaalenii_PYR-1 LGDRAEHRPSELSGGQMQRVAVARALMMDPPLILADEPTGNLDTKTGAAI
MAV_4425|M.avium_104 LGNRTEHRPAELSGGQMQRVAVARALMMDPPLILADEPTGNLDSSTGAAI
Mb2593|M.bovis_AF2122/97 LGERMKHRPGDMSGGQQQRVAVARAIALDPQLILADEPTAHLDFIQVEEV
Rv2564|M.tuberculosis_H37Rv LGERMKHRPGDMSGGQQQRVAVARAIALDPQLILADEPTAHLDFIQVEEV
Mflv_4127|M.gilvum_PYR-GCK LGHRINHQPTRMSGGEQQRTAIARSLISNPALVLADEPTANLDHRTGETV
MSMEG_2524|M.smegmatis_MC2_155 LGHRIHHQPTKMSGGEQQRTAIARSLISNPTLVLADEPTANLDHRTGETV
MAB_4671c|M.abscessus_ATCC_199 MEHRTDHRPPALSGGEKQRVAAARALVARPGLVLADEPTANLDSATGKRV
MMAR_0922|M.marinum_M LADRAGSFPGQLSGGQQQRVAIARALITDPDLLLADEPMGALDVATGRRV
MUL_0674|M.ulcerans_Agy99 LADRAGSFPGQLSGGQQQRVAIARALITNPDLLLADEPMGALDVATGRRV
MLBr_00669|M.leprae_Br4923 LSGKANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDI
TH_4101|M.thermoresistible__bu LADKARRVPAELSGGEQQRVAIARALFMRPDVILADEPTGNLDTKNGRIV
: : * :***: **.* **:: * ::***** . ** :
Mvan_1004|M.vanbaalenii_PYR-1 MALLTDIAHQD--ERSVVMVTHNLGAASAT-DRVITLT------------
MAV_4425|M.avium_104 LELLAEVAHEGGGDRLVVMVTHNSDAAAAT-DRVITLQ------------
Mb2593|M.bovis_AF2122/97 LRLIRSLAQGD---RVVVVATHDSRMLPLA-DRVLELMPAQVSPNQPPET
Rv2564|M.tuberculosis_H37Rv LRLIRSLAQGD---RVVVVATHDSRMLPLA-DRVLELMPAQVSPNQPPET
Mflv_4127|M.gilvum_PYR-GCK VRMLRDLCSTM--GVTVVASTHDPTVADEA-SRVVRMK------------
MSMEG_2524|M.smegmatis_MC2_155 VRMLRDLCLTL--GVTVVASTHDPTVADEA-SRVVRMR------------
MAB_4671c|M.abscessus_ATCC_199 AAQLADAARSQ--GAAVVIVTHDLRLTDIA-DRVLHLE------------
MMAR_0922|M.marinum_M LELLQQTNRSG---LGVIMVTHNRAAADVA-DRIIQMR------------
MUL_0674|M.ulcerans_Agy99 LELLQQTNRSG---LGVIMVTHNRAAADVA-DPIIQMR------------
MLBr_00669|M.leprae_Br4923 MDLLERINRTG---TTVLMATHDHHIVDSMRQRVVELS------------
TH_4101|M.thermoresistible__bu LDTLKKLNAAG---QTIVMVTHDLSIAEEA-PRLIAFR------------
: :: **: :: :
Mvan_1004|M.vanbaalenii_PYR-1 ----------------------------------DG--------------
MAV_4425|M.avium_104 ----------------------------------DG--------------
Mb2593|M.bovis_AF2122/97 VHVKAGEVLFEQSTMGDLIYVVSEGEFEIVRELADGGEELVKTAAPGDYF
Rv2564|M.tuberculosis_H37Rv VHVKAGEVLFEQSTMGDLIYVVSEGEFEIVRELADGGEELVKTAAPGDYF
Mflv_4127|M.gilvum_PYR-GCK ----------------------------------DG--------------
MSMEG_2524|M.smegmatis_MC2_155 ----------------------------------DG--------------
MAB_4671c|M.abscessus_ATCC_199 ----------------------------------DG--------------
MMAR_0922|M.marinum_M ----------------------------------DG--------------
MUL_0674|M.ulcerans_Agy99 ----------------------------------DG--------------
MLBr_00669|M.leprae_Br4923 ----------------------------------LG--------------
TH_4101|M.thermoresistible__bu ----------------------------------DG--------------
*
Mvan_1004|M.vanbaalenii_PYR-1 ---------------RIGSDVLAGGELT----------------------
MAV_4425|M.avium_104 ---------------RVGSDVLAVAG------------------------
Mb2593|M.bovis_AF2122/97 GEIGVLFHLPRSATVRARSDATAVGYTAQAFRERLGVTRVADLIEHRELA
Rv2564|M.tuberculosis_H37Rv GEIGVLFHLPRSATVRARSDATAVGYTAQAFRERLGVTRVADLIEHRELA
Mflv_4127|M.gilvum_PYR-GCK -------------------QIVN---------------------------
MSMEG_2524|M.smegmatis_MC2_155 -------------------QIIN---------------------------
MAB_4671c|M.abscessus_ATCC_199 -------------------VLLEKENQ-----------------------
MMAR_0922|M.marinum_M -------------------AIVEQRRNSAPAEASSVRW------------
MUL_0674|M.ulcerans_Agy99 -------------------AIVEQRRNSAPAEASSVRW------------
MLBr_00669|M.leprae_Br4923 -------------------RLVRD---------------EWCGIYGMDR-
TH_4101|M.thermoresistible__bu -------------------RIESDSAVPASGAAAETQSAEPDGVVVMLRK
Mvan_1004|M.vanbaalenii_PYR-1 ---
MAV_4425|M.avium_104 ---
Mb2593|M.bovis_AF2122/97 SE-
Rv2564|M.tuberculosis_H37Rv SE-
Mflv_4127|M.gilvum_PYR-GCK ---
MSMEG_2524|M.smegmatis_MC2_155 ---
MAB_4671c|M.abscessus_ATCC_199 ---
MMAR_0922|M.marinum_M ---
MUL_0674|M.ulcerans_Agy99 ---
MLBr_00669|M.leprae_Br4923 ---
TH_4101|M.thermoresistible__bu TGR