For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTTIDSANIDENFRETIDGSTGEFKEMYLKSSAALQQALRDNKASAHGVVVESAIKSTSPDTVVVVLFVD QSVSNARAPEPRLDRSRVEVTMKKVDGRWLAAKVELP
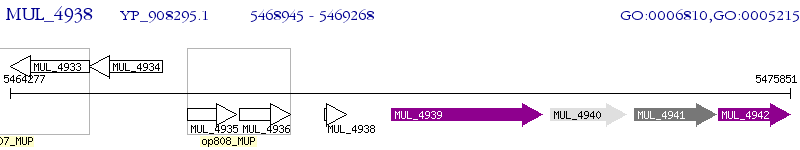
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4938 | - | - | 100% (107) | hypothetical protein MUL_4938 |
| M. ulcerans Agy99 | MUL_3802 | - | 6e-37 | 63.21% (106) | hypothetical protein MUL_3802 |
| M. ulcerans Agy99 | MUL_4935 | - | 2e-35 | 62.26% (106) | hypothetical protein MUL_4935 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2411c | - | 3e-31 | 55.66% (106) | hypothetical protein Mb2411c |
| M. gilvum PYR-GCK | Mflv_0004 | - | 2e-35 | 64.15% (106) | hypothetical protein Mflv_0004 |
| M. tuberculosis H37Rv | Rv2390c | - | 3e-31 | 55.66% (106) | hypothetical protein Rv2390c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4593c | - | 2e-31 | 55.66% (106) | hypothetical protein MAB_4593c |
| M. marinum M | MMAR_0159 | - | 7e-55 | 99.07% (107) | hypothetical protein MMAR_0159 |
| M. avium 104 | MAV_0098 | - | 4e-38 | 68.22% (107) | hypothetical protein MAV_0098 |
| M. smegmatis MC2 155 | MSMEG_1149 | - | 1e-34 | 60.75% (107) | hypothetical protein MSMEG_1149 |
| M. thermoresistible (build 8) | TH_0161 | - | 2e-35 | 62.62% (107) | conserved hypothetical protein |
| M. vanbaalenii PYR-1 | Mvan_0965 | - | 2e-38 | 69.16% (107) | hypothetical protein Mvan_0965 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4938|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0159|M.marinum_M MEGKDDGCISPQEGGTTRKPRRVGPDAKAKAGRTSATTDAAAQPGPDPTR
Mflv_0004|M.gilvum_PYR-GCK ---------------------------------MTVTTQSTRDDEPDTSA
MSMEG_1149|M.smegmatis_MC2_155 ---------------------------------MKLKTDKRRD-VFDEAV
Mvan_0965|M.vanbaalenii_PYR-1 ---------------------------------MSVTTDRDLD---DEKT
TH_0161|M.thermoresistible__bu ---------------------------------VTLTDEDRDRTDRDETP
Mb2411c|M.bovis_AF2122/97 ---------------------------------MAIFG----------RG
Rv2390c|M.tuberculosis_H37Rv ---------------------------------MAIFG----------RG
MAV_0098|M.avium_104 ---------------------------------MQLMNDRKPLQTVGNRG
MAB_4593c|M.abscessus_ATCC_199 ---------------------------------MADDAADKDAKNADESA
MUL_4938|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0159|M.marinum_M DPVSDSPETCGGGHGGNTIGRAPGTAEPSSQETELEVDHDDLDPGIQACA
Mflv_0004|M.gilvum_PYR-GCK DDEQTPEGAGPGDAD----------VADAHTETE----------------
MSMEG_1149|M.smegmatis_MC2_155 DDTEDAEVEALDVAE----------SEDTATEPESDEVTGPSGE------
Mvan_0965|M.vanbaalenii_PYR-1 DGEEDTTSQEEPDG-----------TEDSAAEDQ----------------
TH_0161|M.thermoresistible__bu EPESSEQVVTETSAE----------EAGETGETG----------------
Mb2411c|M.bovis_AF2122/97 -----------------------------HGASEP---------------
Rv2390c|M.tuberculosis_H37Rv -----------------------------HGASEP---------------
MAV_0098|M.avium_104 TWLVRRFRVAAEGIA----------DDEVAPADEPDVAEASPNQ------
MAB_4593c|M.abscessus_ATCC_199 VTSDAEKAESSAPSSAAVESTAEGTTEAATAEVADFDAEDSEKTKDSGAV
MUL_4938|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0159|M.marinum_M AAGDGPDPALEPGHDVTPQRRARS---WTKVAAALAAAAVLAGTAALGWL
Mflv_0004|M.gilvum_PYR-GCK --PADDDSD--PDEPMTARGWRRR--VLLGVVA-LVFAALLAVSGVLGWM
MSMEG_1149|M.smegmatis_MC2_155 -KPSDKPSDEKPAATVTSPNQRRG--VVLGVAAGILVAALLGGGGFVGWQ
Mvan_0965|M.vanbaalenii_PYR-1 -QPEDSASGD-PGPEAGNRSWRRR--VLAGAIAAVTIGAVAT-AGYLGWL
TH_0161|M.thermoresistible__bu -ETGETEKTGEPGEPGPAARSRRAGWLRVGAVT-AVFVVLLGLCGFLGWQ
Mb2411c|M.bovis_AF2122/97 --GGTGEPAETPG---RGRLTR----SVTGWVGAVAVVVSLAGSGWCGWV
Rv2390c|M.tuberculosis_H37Rv --GGTGEPAETPG---RGRLTR----SVIGWVGAVAVVVSLAGSGWCGWV
MAV_0098|M.avium_104 -SEQTDKAAEGEGEPPRPRKRRRI--KLVRAIAAGILAAVLATAGYEGWL
MAB_4593c|M.abscessus_ATCC_199 EDSDDEDISLEPDEEENPKRIRGR--HWALTAAAIVAIAIVALAAIIGRL
MUL_4938|M.ulcerans_Agy99 ------------------------MTTIDSANIDENFRETIDGSTGEFKE
MMAR_0159|M.marinum_M WKEQRDTEIAGRQALSAALRFTENLTTIDSANIDENFRETIDGSTGEFKE
Mflv_0004|M.gilvum_PYR-GCK TWKQHQLTQAGEQARQAAVTYAQVLTSIDSAKVDENFNQVLDGATGEFKD
MSMEG_1149|M.smegmatis_MC2_155 LWQTKQVIAAGNQARDAAVNYAQVLTSIDSNKVDENFDQVLAGATGEFKD
Mvan_0965|M.vanbaalenii_PYR-1 LWQQNQVSRAGEQAQAAAVQYAQILTSIDSAKVDENFDQVLDGATGEFKD
TH_0161|M.thermoresistible__bu VWQQRQLDRAADEARQAAVEYARILTSIDADKVDENFAQVLDGATGEFRD
Mb2411c|M.bovis_AF2122/97 LFEKHQTDVAAGQALQAARSYVVKLATMDCERIDHNMRDILEGSTGEFKD
Rv2390c|M.tuberculosis_H37Rv LFEKHQTDVAAGQALQAARSYVVKLATMDCERIDHNMRDILEGSTGEFKD
MAV_0098|M.avium_104 LFEQHQKDVAAAQALDAAKKYILALTSVDTNAIDKNFAEVLDGSTGEFKD
MAB_4593c|M.abscessus_ATCC_199 YEEQRIEEQSSREALAAAQEFANVMINVDSAKLDESSNKTLERSTGDFKQ
: .:* :*.. . : :**:*::
MUL_4938|M.ulcerans_Agy99 MYLKSSAALQQALRDNKASAHGVVVESAIKSTSPDTVVVVLFVDQSVSNA
MMAR_0159|M.marinum_M MYLKSSAALQQALRENKASAHGVVVESAIKSTSPDTVVVVLFVDQSVSNA
Mflv_0004|M.gilvum_PYR-GCK MYSESSMQLRQLLIDNKAASRGVVVESAIQSVEEGRVVVLLFVDQSVTNS
MSMEG_1149|M.smegmatis_MC2_155 MYSQSSMQLRQLLIENKATAHGVVVESAVQSATKNKAVVLLFVDQSVQNT
Mvan_0965|M.vanbaalenii_PYR-1 MYSQSSVQLRQLLIDNKASAHGVVVDSAIQSESKDKVVVLLFVDQSVSNT
TH_0161|M.thermoresistible__bu MYSASSAQLRQLLIDNKASARGVVLESAVQSAGEDEVVVLLFIDQTVSNA
Mb2411c|M.bovis_AF2122/97 KYGKSSAHLRQLLADNRVATHGTVVAASVKSATTNKVVVLMFIDQSVSNR
Rv2390c|M.tuberculosis_H37Rv KYGKSSAHLRQLLADNRVATHGTVVAASVKSATTNKVVVLMFIDQSVSNR
MAV_0098|M.avium_104 MYTKSSAQLRQTLIDNKAAAHGNVVDAAVRSATEDKVEVVLFVDQSVSNG
MAB_4593c|M.abscessus_ATCC_199 KYSESSTTLRQVMIENKAKATGVVVDSAVKSAAPDKVEVLLFIDQAVSNV
* ** *:* : :*:. : * *: ::::* . . *::*:**:* *
MUL_4938|M.ulcerans_Agy99 RAPEPRLDRSRVEVTMKKVDGRWLAAKVELP
MMAR_0159|M.marinum_M RAPEPRLDRSRVEVTMKKVDGRWLAAKVELP
Mflv_0004|M.gilvum_PYR-GCK AVPDPRIDRSRIKMTMEYVDGRWRAAKVELA
MSMEG_1149|M.smegmatis_MC2_155 AVPDPRIDRSRIKMTMEKVDGQWKASKVELP
Mvan_0965|M.vanbaalenii_PYR-1 SVPDPRIDRSRVKMTMEYVDGRWRASKVELP
TH_0161|M.thermoresistible__bu HVPEPRIDRSRVKMTMRHVDGTWRASKVALP
Mb2411c|M.bovis_AF2122/97 NSPTPQIDRSRIKVIMDKVNGRWLASKVELL
Rv2390c|M.tuberculosis_H37Rv NSPTPQIDRSRIKVIMDKVNGRWLASKVELL
MAV_0098|M.avium_104 AAPAPQLDRSRVKMTMEKVDGRWLASKVELP
MAB_4593c|M.abscessus_ATCC_199 AIPDARMDRSRVQITMQKVDGRWLASDVELT
* .::****::: * *:* * *:.* *