For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QLMNDRKPLQTVGNRGTWLVRRFRVAAEGIADDEVAPADEPDVAEASPNQSEQTDKAAEGEGEPPRPRKR RRIKLVRAIAAGILAAVLATAGYEGWLLFEQHQKDVAAAQALDAAKKYILALTSVDTNAIDKNFAEVLDG STGEFKDMYTKSSAQLRQTLIDNKAAAHGNVVDAAVRSATEDKVEVVLFVDQSVSNGAAPAPQLDRSRVK MTMEKVDGRWLASKVELP*
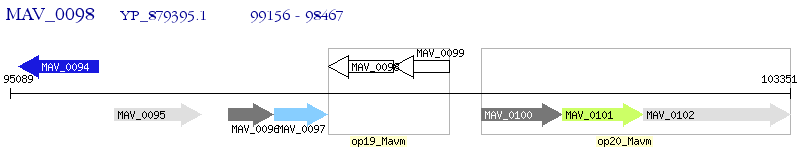
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0098 | - | - | 100% (229) | hypothetical protein MAV_0098 |
| M. avium 104 | MAV_0108 | - | 2e-42 | 46.46% (198) | hypothetical protein MAV_0108 |
| M. avium 104 | MAV_0938 | - | 5e-18 | 27.75% (209) | hypothetical protein MAV_0938 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2411c | - | 7e-48 | 55.88% (170) | hypothetical protein Mb2411c |
| M. gilvum PYR-GCK | Mflv_0004 | - | 2e-48 | 47.64% (212) | hypothetical protein Mflv_0004 |
| M. tuberculosis H37Rv | Rv2390c | - | 4e-48 | 54.71% (170) | hypothetical protein Rv2390c |
| M. leprae Br4923 | MLBr_02615 | - | 8e-07 | 29.11% (158) | hypothetical protein MLBr_02615 |
| M. abscessus ATCC 19977 | MAB_1004c | - | 6e-41 | 42.86% (196) | hypothetical protein MAB_1004c |
| M. marinum M | MMAR_3870 | - | 4e-49 | 53.68% (190) | hypothetical protein MMAR_3870 |
| M. smegmatis MC2 155 | MSMEG_1149 | - | 4e-51 | 50.45% (224) | hypothetical protein MSMEG_1149 |
| M. thermoresistible (build 8) | TH_0161 | - | 1e-46 | 50.00% (200) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3802 | - | 1e-48 | 53.16% (190) | hypothetical protein MUL_3802 |
| M. vanbaalenii PYR-1 | Mvan_0965 | - | 3e-54 | 52.79% (197) | hypothetical protein Mvan_0965 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2411c|M.bovis_AF2122/97 --------------------------------------------------
Rv2390c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1149|M.smegmatis_MC2_155 ---------MKLKTDKRRDVFDEAVDDTEDAEVEALDVAESEDTATEPES
TH_0161|M.thermoresistible__bu ---------VTLTDEDR-----DRTDRDETPEPESSEQVVTETSAEEAGE
Mflv_0004|M.gilvum_PYR-GCK ---------MTVTTQST---------RDDEPDTSADDEQTPEG--AGPGD
Mvan_0965|M.vanbaalenii_PYR-1 ---------MSVTTDRD----------LDDEKTDGEEDTTSQE---EPDG
MMAR_3870|M.marinum_M MHVQRKEMIVVATTEKD------DETQSEEPAASSEDNQDTEDTSQTPDI
MUL_3802|M.ulcerans_Agy99 -------MIVVATTEKD------DETQSEEPAASSEDNQDTEDTSQTPDI
MAB_1004c|M.abscessus_ATCC_199 ---------MVTMTKED-------VDSTNDADTDTADDKETVDSAAETDS
MAV_0098|M.avium_104 ---------MQLMNDRK------PLQTVGNRGTWLVRRFRVAAEGIADDE
MLBr_02615|M.leprae_Br4923 -----------MRNRWR--------LLAFDVVAPLVAIAALVMIGVVLDW
Mb2411c|M.bovis_AF2122/97 -----MAIFGRGHGASEP------GGTGEPAETPGRGRLTRSVTGWVGAV
Rv2390c|M.tuberculosis_H37Rv -----MAIFGRGHGASEP------GGTGEPAETPGRGRLTRSVIGWVGAV
MSMEG_1149|M.smegmatis_MC2_155 D--EVTGPSGE-KPSDKP------SDEKPAATVTSPNQRRGVVLGVAAGI
TH_0161|M.thermoresistible__bu T--GETGETGETEKTGEP------GEPGPAARSRRAGWLR---VGAVTAV
Mflv_0004|M.gilvum_PYR-GCK A--DVADAHTETEPADDD------SDPDEPMTAR--GWRRRVLLGVVALV
Mvan_0965|M.vanbaalenii_PYR-1 T--EDSAAEDQQPEDSAS------GDPGPEAGNR--SWRRRVLAGAIAAV
MMAR_3870|M.marinum_M E--VDSVEHAEAESADDDEDNTLLADKREKTARPNRSWARYLRRGAMPLL
MUL_3802|M.ulcerans_Agy99 E--VDSVEHAEAESADDDEDNTLLADKREKAARPNRSWARYLRRGAMPLL
MAB_1004c|M.abscessus_ATCC_199 EIAEDAASTDTDAASSADSDSETASSDEDGDAAKGASWVRYAALGAVAAI
MAV_0098|M.avium_104 VAPADEPDVAEASPNQSEQ--TDKAAEGEGEPPRPRKRRRIKLVRAIAAG
MLBr_02615|M.leprae_Br4923 PRWWVSACSVLVLLIVEGVGVNFWLLRRDSVTIGTDDDAPGLRLAVVSVC
Mb2411c|M.bovis_AF2122/97 AVVVSLAGSGWCGWVLFE--KHQTDVAAGQALQAARSYVVKLATMD----
Rv2390c|M.tuberculosis_H37Rv AVVVSLAGSGWCGWVLFE--KHQTDVAAGQALQAARSYVVKLATMD----
MSMEG_1149|M.smegmatis_MC2_155 LVAALLGGGGFVGWQLWQ--TKQVIAAGNQARDAAVNYAQVLTSID----
TH_0161|M.thermoresistible__bu FVV-LLGLCGFLGWQVWQ--QRQLDRAADEARQAAVEYARILTSID----
Mflv_0004|M.gilvum_PYR-GCK FAA-LLAVSGVLGWMTWK--QHQLTQAGEQARQAAVTYAQVLTSID----
Mvan_0965|M.vanbaalenii_PYR-1 TIG-AVATAGYLGWLLWQ--QNQVSRAGEQAQAAAVQYAQILTSID----
MMAR_3870|M.marinum_M LAG-ALALSAFLGWQQWQ--QHQVKLAGQQAQQAAIAYAQVLTSID----
MUL_3802|M.ulcerans_Agy99 LAG-ALALSAFLDWQQWQ--QHQVKLAGQQAQQAAIAYAQVLTSID----
MAB_1004c|M.abscessus_ATCC_199 MVG-ALVFSGIFGWKVWQ--QHQIDAAGEQALAVAIPYTETLTSID----
MAV_0098|M.avium_104 ILAAVLATAGYEGWLLFE--QHQKDVAAAQALDAAKKYILALTSVD----
MLBr_02615|M.leprae_Br4923 TAA--LCAAVLIGYMHWTSPDRDFSLDSREVVQIATGMAEAMASFTPSAP
: .: : .: . :. * :::.
Mb2411c|M.bovis_AF2122/97 CERIDHNMRDILEGSTGEFKDKYGKSSAHLRQLLADNRVATHGTVVAASV
Rv2390c|M.tuberculosis_H37Rv CERIDHNMRDILEGSTGEFKDKYGKSSAHLRQLLADNRVATHGTVVAASV
MSMEG_1149|M.smegmatis_MC2_155 SNKVDENFDQVLAGATGEFKDMYSQSSMQLRQLLIENKATAHGVVVESAV
TH_0161|M.thermoresistible__bu ADKVDENFAQVLDGATGEFRDMYSASSAQLRQLLIDNKASARGVVLESAV
Mflv_0004|M.gilvum_PYR-GCK SAKVDENFNQVLDGATGEFKDMYSESSMQLRQLLIDNKAASRGVVVESAI
Mvan_0965|M.vanbaalenii_PYR-1 SAKVDENFDQVLDGATGEFKDMYSQSSVQLRQLLIDNKASAHGVVVDSAI
MMAR_3870|M.marinum_M SNNVDQNFRQVLDGATGEFKDMYTQSSVQLRQLLIDNKASAHGVVVDSAV
MUL_3802|M.ulcerans_Agy99 SNNVDQNFRQVLDGATGEFKDMYTQSSVQLRQLLIDNKASAHGVVVDSAV
MAB_1004c|M.abscessus_ATCC_199 SNDIDESFRKILNGATGDLKKDFTPASVQLRQLLLDNKAASHGKVVNSAI
MAV_0098|M.avium_104 TNAIDKNFAEVLDGSTGEFKDMYTKSSAQLRQTLIDNKAAAHGNVVDAAV
MLBr_02615|M.leprae_Br4923 TSSIDRAAAMIMPDQAGVFKEQYRKSSADLAR----RNVTAQASVLAAGV
:*. :: . :* ::. : :* .* : ...:::. *: :.:
Mb2411c|M.bovis_AF2122/97 KSATTNKVVVLMFIDQSVSNRNSPTPQIDRSRIKVIMDKVNGRWLASKVE
Rv2390c|M.tuberculosis_H37Rv KSATTNKVVVLMFIDQSVSNRNSPTPQIDRSRIKVIMDKVNGRWLASKVE
MSMEG_1149|M.smegmatis_MC2_155 QSATKNKAVVLLFVDQSVQNTAVPDPRIDRSRIKMTMEKVDGQWKASKVE
TH_0161|M.thermoresistible__bu QSAGEDEVVVLLFIDQTVSNAHVPEPRIDRSRVKMTMRHVDGTWRASKVA
Mflv_0004|M.gilvum_PYR-GCK QSVEEGRVVVLLFVDQSVTNSAVPDPRIDRSRIKMTMEYVDGRWRAAKVE
Mvan_0965|M.vanbaalenii_PYR-1 QSESKDKVVVLLFVDQSVSNTSVPDPRIDRSRVKMTMEYVDGRWRASKVE
MMAR_3870|M.marinum_M ASETVNKVVVLVFVDQTVTNMAVPDPRIDRSRIKMTMEKVDGRWLASKVQ
MUL_3802|M.ulcerans_Agy99 ASETVNKVVVLIFVDQTVTNMAVPDPRIDRSRIKMTMEKVDGRWLASKVQ
MAB_1004c|M.abscessus_ATCC_199 QYKSTNKVVLLMMVDQSITNTQRPDPRVDKSRMKITMEKIDGRWLASKLE
MAV_0098|M.avium_104 RSATEDKVEVVLFVDQSVSNGAAPAPQLDRSRVKMTMEKVDGRWLASKVE
MLBr_02615|M.leprae_Br4923 EAIGSSAASVAVILRVTQNTPGQP-PSQAAPAVRVTLIKRGSDWLVTDVL
. . : ::: : . * * . ::: : .. * .:.:
Mb2411c|M.bovis_AF2122/97 LL---
Rv2390c|M.tuberculosis_H37Rv LL---
MSMEG_1149|M.smegmatis_MC2_155 LP---
TH_0161|M.thermoresistible__bu LP---
Mflv_0004|M.gilvum_PYR-GCK LA---
Mvan_0965|M.vanbaalenii_PYR-1 LP---
MMAR_3870|M.marinum_M LL---
MUL_3802|M.ulcerans_Agy99 LL---
MAB_1004c|M.abscessus_ATCC_199 YL---
MAV_0098|M.avium_104 LP---
MLBr_02615|M.leprae_Br4923 SINAR