For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LQDKVVVVTGGSRGLGRAMVRAFAAHGADVVIASRKIDSCQELAAQVEAEHGRRALPVACNVSSWEQCDH LVDTVYREFGRVDVLVNNAGLSPLYPSLDQVSEALFDKVIGVNLKGPFRLSASIATKMAAGAGGSIINIS SIEAVRPDATALPYAAAKAGLNALTLGLSHTFGPTVRTNTIQCGLFNTDIAAAWPEGFAEALIPSIPLRR VGEPEDIVGAALYLASEASAYCTGTTIRLDGGIL*
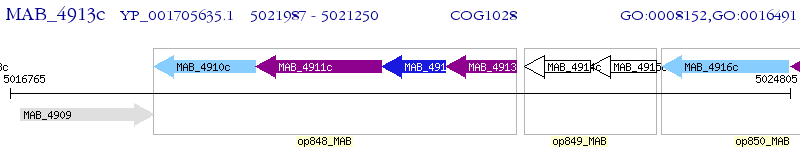
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4913c | - | - | 100% (245) | putative short chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_0833 | - | 1e-33 | 34.29% (245) | Short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_2032 | - | 4e-33 | 35.37% (246) | 3-oxoacyl- |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2788c | fabG | 2e-37 | 35.66% (244) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1202 | - | 4e-69 | 53.31% (242) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2766c | fabG | 2e-37 | 35.66% (244) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 4e-25 | 31.80% (239) | 3-oxoacyl- |
| M. marinum M | MMAR_5310 | - | 2e-71 | 54.51% (244) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0311 | - | 4e-69 | 52.87% (244) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_6340 | - | 2e-71 | 54.73% (243) | short-chain dehydrogenase/reductase SDR |
| M. thermoresistible (build 8) | TH_3170 | - | 3e-78 | 58.16% (239) | PUTATIVE short-chain type dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_4384 | - | 3e-71 | 54.51% (244) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_5605 | - | 4e-69 | 55.10% (245) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2788c|M.bovis_AF2122/97 -------------MTSLDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVL
Rv2766c|M.tuberculosis_H37Rv -------------MTSLDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVL
MMAR_5310|M.marinum_M ---------MGYADQLFDLTDHVVLITGGSRGLGRQMAFAAARCGADVVI
MUL_4384|M.ulcerans_Agy99 ---------MGYADQLFDLTDHVVLITGGSRGLGRQMAFAAARCGANVVI
MAV_0311|M.avium_104 ---------MGYAEQLFDLADRVVLITGGSRGLGREMAFGAARCGADVVI
MSMEG_6340|M.smegmatis_MC2_155 ---------MGYADQLFDLTDRVVLITGGSRGLGREMAFGAARCGADVII
Mflv_1202|M.gilvum_PYR-GCK -----MT---GFADRLFDLTDRVVLITGGSRGLGREMAVAAAHCGADVII
Mvan_5605|M.vanbaalenii_PYR-1 -----MTASGGFADQLFDLTDRVVLVTGGSRGLGREMAFAAARCGADVVI
TH_3170|M.thermoresistible__bu -----------MENPNLSMAGKVALVTGGSRGLGRAMTLGFARAGADVII
MAB_4913c|M.abscessus_ATCC_199 -----------------MLQDKVVVVTGGSRGLGRAMVRAFAAHGADVVI
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
: .: ::**..**:* :. . * * :
Mb2788c|M.bovis_AF2122/97 TARRQE----AADEAAAQVGDRALGVGAHAVDEDAARRCVDLTLERFGSV
Rv2766c|M.tuberculosis_H37Rv TARRQE----AADEAAAQVGDRALGVGAHAVDEDAARRCVDLTLERFGSV
MMAR_5310|M.marinum_M ASRNLDNCVATATEIESETGRCAMAYQVHVGRWDQLDGLVAASYERFGKI
MUL_4384|M.ulcerans_Agy99 ASRNLDNCVATATEIESETGRSALAYQVHVGRWDQLDGLVAASYERFGKI
MAV_0311|M.avium_104 ASRNLDNCVATAQQIEHETGRRAMAYQVHVGRWDQLDGLVEASYDRFGKI
MSMEG_6340|M.smegmatis_MC2_155 ASRNLDSCVVTAEEIASETGRTAFPYSVHVGHWDELDGLVDAAYERFGKV
Mflv_1202|M.gilvum_PYR-GCK ASRKYDACVATAEEITAQTGRAALPYGVHVGRWDELDGLVDAAYARFGKV
Mvan_5605|M.vanbaalenii_PYR-1 ASRKYEACVATADEIAAETGRAAFPYQVHVGRWDELDGLVDAAYDRFGRV
TH_3170|M.thermoresistible__bu ASRKFDSCKELATEIEQTIGRRAVPIAANVGNWEDCDALYRAAYEAFPRV
MAB_4913c|M.abscessus_ATCC_199 ASRKIDSCQELAAQVEAEHGRRALPVACNVSSWEQCDHLVDTVYREFGRV
MLBr_01807|M.leprae_Br4923 THR------------ASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQGPV
: * . .. : :
Mb2788c|M.bovis_AF2122/97 DILINNAGTNPAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEH
Rv2766c|M.tuberculosis_H37Rv DILINNAGTNPAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEH
MMAR_5310|M.marinum_M DTLINNAGMSPLYDKLSDVTEKLFDAVLNLNLKGPFRLSALVGERMVAAG
MUL_4384|M.ulcerans_Agy99 DTLINNAGMSPLYDKLSDVTEKLFDAVLNLNLKGPFRLSALVGERMVAAD
MAV_0311|M.avium_104 DTLINNAGMSPLYDKLTDVTEKLFDAVVNLNLKGPFRLSALVGERMVAAG
MSMEG_6340|M.smegmatis_MC2_155 DVLVNNAGMSPLYESLSAVTEKLYDAVFNLNLKGPFRLSALIGERMVADG
Mflv_1202|M.gilvum_PYR-GCK DVLVNNAGMSPVYDKQSDVTEKLFDAVVNLNLKGPFRLSALIGERMVADG
Mvan_5605|M.vanbaalenii_PYR-1 DVLVNNAGMSPLYESLSAVTEKLFDSVVNLNLKGPFRLSVLVGDRMVADG
TH_3170|M.thermoresistible__bu DVLVNNAGMSPLYDKPSDVTEALYDKVLGVNLRGPFRLTALFGERMQADG
MAB_4913c|M.abscessus_ATCC_199 DVLVNNAGLSPLYPSLDQVSEALFDKVIGVNLKGPFRLSASIATKMAAGA
MLBr_01807|M.leprae_Br4923 EVLVANAGITSDMLIMR-MSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR
: *: *** .. . : :..:** . : :
Mb2788c|M.bovis_AF2122/97 GGAVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSP-RIRVNAI
Rv2766c|M.tuberculosis_H37Rv GGAVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSP-RIRVNAI
MMAR_5310|M.marinum_M GGSIINVSSAGSLRPSADIIPYAAAKAGLNAMTEGLARAFGP-TVRVNTL
MUL_4384|M.ulcerans_Agy99 GGSIINVSSAGSLRPSADIIPYAAAKAGLNAMTEGLARAFGP-TVRVNTL
MAV_0311|M.avium_104 RGSIINVSTAGSLRPTPDIVPYAASKAGLNAMTEALAKAFGP-AVRVNTL
MSMEG_6340|M.smegmatis_MC2_155 GGRIINVSSSGSLRPDQYMLPYAAAKAGLNVLTEGLAKAYGP-TVRVNTL
Mflv_1202|M.gilvum_PYR-GCK GGSIVNVSTHGSLRPHPSFIPYAASKAGLNAMTEGLALAFGP-SVRVNTL
Mvan_5605|M.vanbaalenii_PYR-1 GGSIINVSTHGSIRPHPSFVPYAASKAGLNAMTEALALAYGP-TVRVNTL
TH_3170|M.thermoresistible__bu GGSIINISSVSGVNPGANVIPYGAAKAGLNSITVSFARAYGP-RVRVNCI
MAB_4913c|M.abscessus_ATCC_199 GGSIINISSIEAVRPDATALPYAAAKAGLNALTLGLSHTFGP-TVRTNTI
MLBr_01807|M.leprae_Br4923 FGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITANVV
* : .: .: * *:*:.* :: :: .. : .* :
Mb2788c|M.bovis_AF2122/97 CPGVVRTRLAEALWKDHEDP-LAATIALGRIGEPADIASAVAFLVSDAAS
Rv2766c|M.tuberculosis_H37Rv CPGVVRTRLAEALWKDHEDP-LAATIALGRIGEPADIASAVAFLVSDAAS
MMAR_5310|M.marinum_M MAGPFLTDVSKAWNLDAATQNPFGHLTLRRAGNPPEIVGAALFLASDASS
MUL_4384|M.ulcerans_Agy99 MAGPFLTDVSKAWNLDAATQNPFGHLALRRAGNPPEIVGAALFLASDASS
MAV_0311|M.avium_104 MAGPFLTDVSRAWNLEAAQENPFRHLALQRAGDPREIVGAALFLASDASS
MSMEG_6340|M.smegmatis_MC2_155 MAGPFLTDVSRAWNMSGD---PFSHLALRRAGDPREIVGAALFLMSDASS
Mflv_1202|M.gilvum_PYR-GCK MPGPFLTDISNAWNF-EGTDNPFRGSALQRAGQPAEIVGAALFLMSDASS
Mvan_5605|M.vanbaalenii_PYR-1 MPGPFLTDISDAWTFTDGDTNPFGHSALQRAGQPAEIVGAALFLMSDASS
TH_3170|M.thermoresistible__bu MVGRFRTDVAKYWDEEAT-AAEIEQYALRRIGEPDEVVGTALYLATDASS
MAB_4913c|M.abscessus_ATCC_199 QCGLFNTDIAAAWPEGFA-EALIPSIPLRRVGEPEDIVGAALYLASEASA
MLBr_01807|M.leprae_Br4923 APGFIDTEMTRAMDSKYQ-EAALQAIPLGRIGAVEEIAGAVSFLASEDAG
* . * :: .* * * ::..:. :* :: :.
Mb2788c|M.bovis_AF2122/97 WITGETMIIDGGLLLGNALGFRAAPSTEH
Rv2766c|M.tuberculosis_H37Rv WITGETMIIDGGLLLGNALGFRAAPSTEH
MMAR_5310|M.marinum_M FTTGSILRADGGIP---------------
MUL_4384|M.ulcerans_Agy99 FTTGSILRADGGIP---------------
MAV_0311|M.avium_104 FTTGSILRADGGIP---------------
MSMEG_6340|M.smegmatis_MC2_155 FTTGSIVRADGGLP---------------
Mflv_1202|M.gilvum_PYR-GCK FTTGSIVRADGGSA---------------
Mvan_5605|M.vanbaalenii_PYR-1 FTTGSIVRADGGIP---------------
TH_3170|M.thermoresistible__bu YTTGAILRVDGGTPY--------------
MAB_4913c|M.abscessus_ATCC_199 YCTGTTIRLDGGIL---------------
MLBr_01807|M.leprae_Br4923 YISGAVIPVDGGLGMGH------------
: :* : ***